4M3N
 
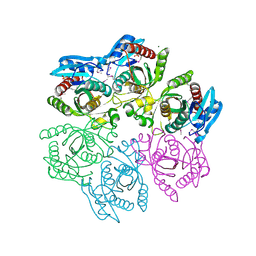 | | Crystal structure of purine nucleoside phosphorylase from Meiothermus ruber DSM 1279, NYSGRC Target 029804. | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-06 | | Release date: | 2013-08-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase from Meiothermus ruber DSM 1279, NYSGRC Target 029804.
To be Published
|
|
4LZA
 
 | | Crystal structure of adenine phosphoribosyltransferase from Thermoanaerobacter pseudethanolicus ATCC 33223, NYSGRC Target 029700. | | Descriptor: | Adenine phosphoribosyltransferase, CHLORIDE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-07-31 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of adenine phosphoribosyltransferase from Thermoanaerobacter pseudethanolicus ATCC 33223, NYSGRC Target 029700.
To be Published
|
|
4MCH
 
 | | Crystal structure of uridine phosphorylase from vibrio fischeri es114 complexed with 6-hydroxy-1-naphthoic acid, NYSGRC Target 029520. | | Descriptor: | 6-hydroxynaphthalene-1-carboxylic acid, DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-21 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of uridine phosphorylase from vibrio fischeri es114 complexed with 6-hydroxy-1-naphthoic acid, NYSGRC Target 029520.
To be Published
|
|
4M1Q
 
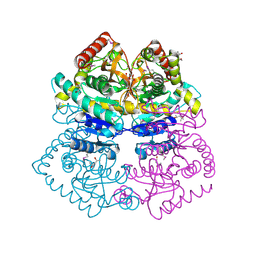 | | Crystal structure of L-lactate dehydrogenase from Bacillus selenitireducens MLS10, NYSGRC Target 029814. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, L-lactate dehydrogenase, PHOSPHATE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-03 | | Release date: | 2013-08-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of L-lactate dehydrogenase from Bacillus selenitireducens MLS10, NYSGRC Target 029814.
To be Published
|
|
4M7W
 
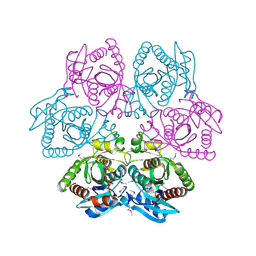 | | Crystal structure of purine nucleoside phosphorylase from Leptotrichia buccalis C-1013-b, NYSGRC Target 029767. | | Descriptor: | PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-12 | | Release date: | 2013-08-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase from Leptotrichia buccalis C-1013-b, NYSGRC Target 029767.
To be Published
|
|
4LYY
 
 | | Crystal structure of hypoxanthine phosphoribosyltransferase from Shewanella pealeana ATCC 700345, NYSGRC Target 029677. | | Descriptor: | Hypoxanthine phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-07-31 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of hypoxanthine phosphoribosyltransferase from Shewanella pealeana ATCC 700345, NYSGRC Target 029677.
To be Published
|
|
4MCI
 
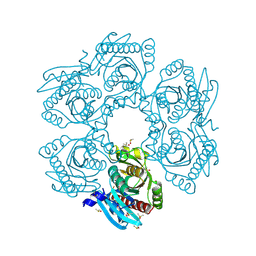 | | Crystal structure of uridine phosphorylase from vibrio fischeri es114 complexed with DMSO, NYSGRC Target 029520. | | Descriptor: | DIMETHYL SULFOXIDE, SULFATE ION, Uridine phosphorylase | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-21 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of uridine phosphorylase from vibrio fischeri es114 complexed with DMSO, NYSGRC Target 029520.
To be Published
|
|
4LNA
 
 | | CRYSTAL STRUCTURE OF purine nucleoside phosphorylase I from Spirosoma linguale DSM 74, NYSGRC Target 029362 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENINE, CHLORIDE ION, ... | | Authors: | Malashkevich, V.N, Bonanno, J.B, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-07-11 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase I from Spirosoma linguale DSM 74, NYSGRC Target 029362
To be Published
|
|
4LNH
 
 | | Crystal structure of uridine phosphorylase from Vibrio fischeri ES114, NYSGRC Target 29520. | | Descriptor: | GLYCEROL, SULFATE ION, Uridine phosphorylase | | Authors: | Malashkevich, V.N, Bonanno, J.B, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-07-11 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of uridine phosphorylase from Vibrio fischeri ES114, NYSGRC Target 29520.
To be Published
|
|
4LCA
 
 | | Crystal structure of probable sugar kinase protein from Rhizobium Etli CFN 42 complexed with thymidine | | Descriptor: | ADENOSINE, DIMETHYL SULFOXIDE, POTASSIUM ION, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-06-21 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium Etli CFN 42 complexed with thymidine
To be Published
|
|
4LBG
 
 | | Crystal structure of probable sugar kinase protein from Rhizobium Etli CFN 42 complexed with adenosine | | Descriptor: | ADENOSINE, DIMETHYL SULFOXIDE, POTASSIUM ION, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-06-20 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium Etli CFN 42 complexed with adenosine
To be Published
|
|
4LC4
 
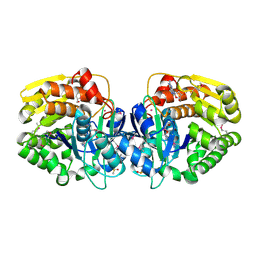 | | Crystal structure of probable sugar kinase protein from Rhizobium Etli CFN 42 complexed with guanosine | | Descriptor: | ADENOSINE, DIMETHYL SULFOXIDE, GUANOSINE, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-06-21 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium Etli CFN 42 complexed with guanosine
To be Published
|
|
4M1E
 
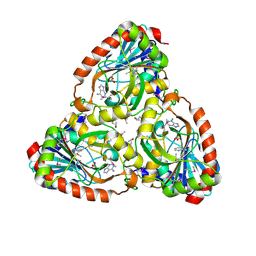 | | Crystal structure of purine nucleoside phosphorylase I from Planctomyces limnophilus DSM 3776, NYSGRC Target 029364. | | Descriptor: | ADENINE, PYRIDINE-2-CARBOXYLIC ACID, Purine nucleoside phosphorylase, ... | | Authors: | Malashkevich, V.N, Bonanno, J.B, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-02 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase I from Planctomyces limnophilus DSM 3776, NYSGRC Target 029364.
To be Published
|
|
4LKR
 
 | | Crystal structure of deoD-3 gene product from Shewanella oneidensis MR-1, NYSGRC target 029437 | | Descriptor: | Purine nucleoside phosphorylase DeoD-type | | Authors: | Malashkevich, V.N, Bonanno, J.B, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-07-08 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of deoD-3 gene product from Shewanella oneidensis MR-1, NYSGRC target 029437
To be Published
|
|
4LMR
 
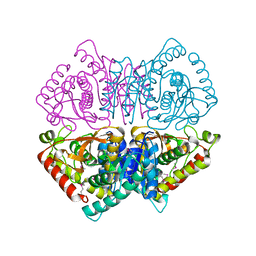 | | Crystal structure of L-lactate dehydrogenase from Bacillus cereus ATCC 14579, NYSGRC Target 029452 | | Descriptor: | L-lactate dehydrogenase 1 | | Authors: | Malashkevich, V.N, Bonanno, J.B, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-07-10 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of L-lactate dehydrogenase from Bacillus cereus ATCC 14579, NYSGRC Target 029452
To be Published
|
|
4LBX
 
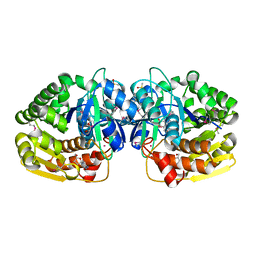 | | Crystal structure of probable sugar kinase protein from Rhizobium Etli CFN 42 complexed with cytidine | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, ADENOSINE, DIMETHYL SULFOXIDE, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-06-21 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium Etli CFN 42 complexed with cytidine
To be Published
|
|
4KAD
 
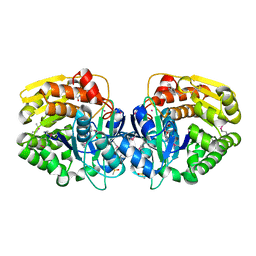 | | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with N1-(2.3-dihydro-1H-inden-5-yl)acetam | | Descriptor: | ADENOSINE, DIMETHYL SULFOXIDE, N-(4,7-dihydro-1H-inden-6-yl)acetamide, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-22 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with N1-(2.3-dihydro-1H-inden-5-yl)acetam
To be Published
|
|
4K9I
 
 | | CRYSTAL STRUCTURE OF probable sugar kinase protein from Rhizobium etli CFN 42 complexed with Norharmane | | Descriptor: | ADENOSINE, DIMETHYL SULFOXIDE, Norharmane, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | CRYSTAL STRUCTURE OF probable sugar kinase protein from Rhizobium etli CFN 42 complexed with Norharmane
To be Published
|
|
4KA0
 
 | | Crystal structure of a putative thiol-disulfide oxidoreductase from Bacteroides vulgatus (target NYSGRC-011676), space group P21221 | | Descriptor: | CHLORIDE ION, Putative thiol-disulfide oxidoreductase | | Authors: | Vetting, M.W, Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R.D, Bonanno, J.B, Armstrong, R.N, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-21 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a putative thiol-disulfide oxidoreductase from Bacteroides vulgatus (target NYSGRC-011676), space group P21221
To be Published
|
|
4K8K
 
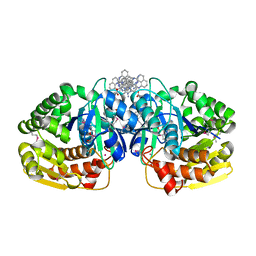 | | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with 1-(4-methoxyphenyl)-1-cyclopropane and 2-aminoperimidine | | Descriptor: | 1-(4-methoxyphenyl)cyclopropanecarboxylic acid, 1H-perimidin-2-amine, ADENOSINE, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-01 | | Last modified: | 2013-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with 1-(4-methoxyphenyl)-1-cyclopropane and 2-aminoperimidine
To be Published
|
|
4M0K
 
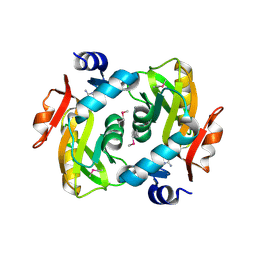 | | Crystal structure of adenine phosphoribosyltransferase from Rhodothermus marinus DSM 4252, NYSGRC Target 029775. | | Descriptor: | ADENOSINE MONOPHOSPHATE, Adenine phosphoribosyltransferase, CALCIUM ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-01 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of adenine phosphoribosyltransferase from Rhodothermus marinus DSM 4252, NYSGRC Target 029775.
To be Published
|
|
4K8C
 
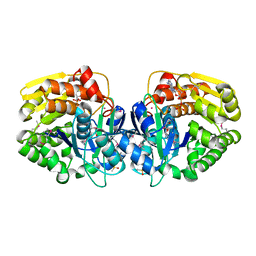 | | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with ADP | | Descriptor: | ADENOSINE, ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with ADP
To be Published
|
|
4K93
 
 | | CRYSTAL STRUCTURE OF probable sugar kinase protein from Rhizobium etli CFN 42 complexed with N-(HYDROXYMETHYL)BENZAMIDE | | Descriptor: | ADENOSINE, DIMETHYL SULFOXIDE, N-(hydroxymethyl)benzamide, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF probable sugar kinase protein from Rhizobium etli CFN 42 complexed with N-(HYDROXYMETHYL)BENZAMIDE
To be Published
|
|
4KBE
 
 | | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with benzoguanamine | | Descriptor: | 6-phenyl-1,3,5-triazine-2,4-diamine, ADENOSINE, DIMETHYL SULFOXIDE, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-23 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with benzoguanamine
To be Published
|
|
4K8T
 
 | | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with ethyl 3,4-diaminobenzoate | | Descriptor: | ADENOSINE, DIMETHYL SULFOXIDE, POTASSIUM ION, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with ethyl 3,4-diaminobenzoate
To be Published
|
|
