6Q4W
 
 | | Structure of MPT-1, a GDP-Man-dependent mannosyltransferase from Leishmania mexicana | | Descriptor: | LmxM MPT-1 | | Authors: | Sobala, L.F, Males, A, Bastidas, L.M, Ward, T, Sernee, M.F, Ralton, J.E, Nero, T.L, Cobbold, S, Kloehn, J, Viera-Lara, M, Stanton, L, Hanssen, E, Parker, M.W, Williams, S.J, McConville, M.J, Davies, G.J. | | Deposit date: | 2018-12-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Family of Dual-Activity Glycosyltransferase-Phosphorylases Mediates Mannogen Turnover and Virulence in Leishmania Parasites.
Cell Host Microbe, 26, 2019
|
|
6Q6W
 
 | | Structure of Fucosylated D-antimicrobial peptide SB5 in complex with the Fucose-binding lectin PA-IIL at 1.438 Angstrom resolution | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, AMINO GROUP, CALCIUM ION, ... | | Authors: | Baeriswyl, S, Stocker, A, Reymond, J.L. | | Deposit date: | 2018-12-12 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.438 Å) | | Cite: | X-ray Crystal Structures of Short Antimicrobial Peptides as Pseudomonas aeruginosa Lectin B Complexes.
Acs Chem.Biol., 14, 2019
|
|
6FLB
 
 | | 3H5 Fab bound to EDIII of DenV 2 Xtal form 2 | | Descriptor: | CHLORIDE ION, Domain III of Dengue virus 2, GLYCEROL, ... | | Authors: | Flanagan, A, Renner, M, Grimes, J.M. | | Deposit date: | 2018-01-25 | | Release date: | 2018-10-24 | | Last modified: | 2018-10-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of a potent and highly unusual minimally enhancing antibody directed against dengue virus.
Nat. Immunol., 19, 2018
|
|
3GCD
 
 | | Structure of the V. cholerae RTX cysteine protease domain in complex with an aza-Leucine peptide inhibitor | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, RTX toxin RtxA, SODIUM ION, ... | | Authors: | Lupardus, P.J, Garcia, K.C, Shen, A, Bogyo, M. | | Deposit date: | 2009-02-21 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanistic and structural insights into the proteolytic activation of Vibrio cholerae MARTX toxin.
Nat.Chem.Biol., 5, 2009
|
|
2BEQ
 
 | | Structure of a Proteolytically Resistant Core from the Severe Acute Respiratory Syndrome Coronavirus S2 Fusion Protein | | Descriptor: | Spike glycoprotein | | Authors: | Supekar, V.M, Bruckmann, C, Ingallinella, P, Bianchi, E, Pessi, A, Carfi, A. | | Deposit date: | 2004-11-29 | | Release date: | 2004-12-22 | | Last modified: | 2020-04-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of a Proteolytically Resistant Core from the Severe Acute Respiratory Syndrome Coronavirus S2 Fusion Protein
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2BEZ
 
 | | Structure of a proteolitically resistant core from the severe acute respiratory syndrome coronavirus S2 fusion protein | | Descriptor: | GLYCEROL, Spike glycoprotein | | Authors: | Supekar, V.M, Bruckmann, C, Ingallinella, P, Bianchi, E, Pessi, A, Carfi, A. | | Deposit date: | 2004-12-02 | | Release date: | 2004-12-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of a Proteolytically Resistant Core from the Severe Acute Respiratory Syndrome Coronavirus S2 Fusion Protein
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6QD5
 
 | | X-ray Structure of the Human Urea Channel SLC14A1/UT1 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, TETRAETHYLENE GLYCOL, Urea transporter 1, ... | | Authors: | Dietz, L, Chi, G, Pike, A.C.W, Moreau, C, Man, H, Snee, M, Scacioc, A, Shrestha, L, Mukhopadhyay, S.M.M, Mckinley, G, Ellis, K, Kliszcak, M, Chalk, R, Borkowska, O, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Durr, K.L, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-31 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | X-ray Structure of the Human Urea Channel SLC14A1/UT1
To Be Published
|
|
2KLT
 
 | | Second Ca2+ binding domain of NCX1.3 | | Descriptor: | Sodium/calcium exchanger 1 | | Authors: | Hilge, M, Aelen, J, Foarce, A, Perrakis, A, Vuister, G.W. | | Deposit date: | 2009-07-08 | | Release date: | 2009-08-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Ca2+ regulation in the Na+/Ca2+ exchanger features a dual electrostatic switch mechanism.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6QEA
 
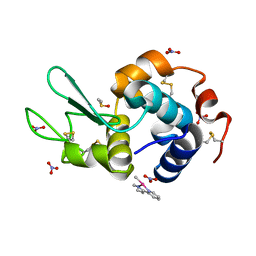 | | The X-ray structure of the adduct formed in the reaction between hen egg white lysozyme and complex I, a pentacoordinate Pt(II) compound containing 2,9-dimethyl-1,10-phenanthroline, dimethylfumarate, methyl and iodine as ligands | | Descriptor: | DIMETHYL SULFOXIDE, Lysozyme C, NITRATE ION, ... | | Authors: | Merlino, A, Ferraro, G. | | Deposit date: | 2019-01-07 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Reaction with Proteins of a Five-Coordinate Platinum(II) Compound.
Int J Mol Sci, 20, 2019
|
|
2BIK
 
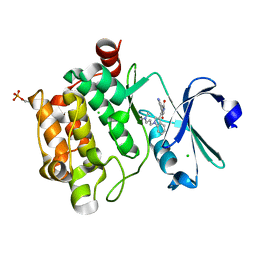 | | Human Pim1 phosphorylated on Ser261 | | Descriptor: | 3-{1-[3-(DIMETHYLAMINO)PROPYL]-1H-INDOL-3-YL}-4-(1H-INDOL-3-YL)-1H-PYRROLE-2,5-DIONE, CHLORIDE ION, PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Knapp, S, Debreczeni, J, Bullock, A, von Delft, F, Sundstrom, M, Arrowsmith, C, Edwards, A, Guo, K. | | Deposit date: | 2005-01-22 | | Release date: | 2005-02-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human Pim1 Phosphorylated on Ser261
To be Published
|
|
3BE5
 
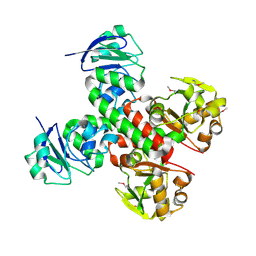 | | Crystal structure of FitE (crystal form 1), a group III periplasmic siderophore binding protein | | Descriptor: | CHLORIDE ION, Putative iron compound-binding protein of ABC transporter family | | Authors: | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2007-11-16 | | Release date: | 2008-10-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trapping open and closed forms of FitE-A group III periplasmic binding protein.
Proteins, 75, 2008
|
|
6FH3
 
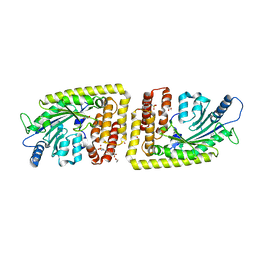 | | Protein arginine kinase McsB in the pArg-bound state | | Descriptor: | 1,2-ETHANEDIOL, Protein-arginine kinase, phospho-arginine | | Authors: | Suskiewicz, M.J, Heuck, A, Vu, L.D, Clausen, T. | | Deposit date: | 2018-01-12 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of McsB, a protein kinase for regulated arginine phosphorylation.
Nat.Chem.Biol., 15, 2019
|
|
6QEV
 
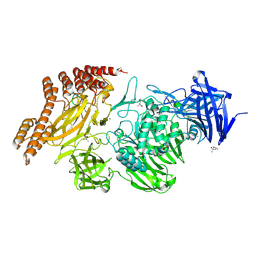 | | EngBF DARPin Fusion 4b B6 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MANGANESE (II) ION, PEGA domain-containing protein,PEGA domain-containing protein,EngBF DARPin fusion B6 complex, ... | | Authors: | Ernst, P, Pluckthun, A, Mittl, P.R.E. | | Deposit date: | 2019-01-08 | | Release date: | 2019-11-06 | | Last modified: | 2020-04-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of biological targets by host:guest crystal lattice engineering.
Sci Rep, 9, 2019
|
|
3G2O
 
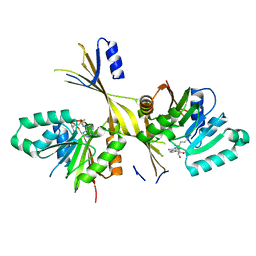 | | Crystal Structure of the Glycopeptide N-methyltransferase MtfA complexed with (S)-adenosyl-L-methionine (SAM) | | Descriptor: | PCZA361.24, S-ADENOSYLMETHIONINE | | Authors: | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2009-01-31 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of the glycopeptide N-methyltransferase MtfA, a tool for the biosynthesis of modified glycopeptide antibiotics.
Chem.Biol., 16, 2009
|
|
6QFK
 
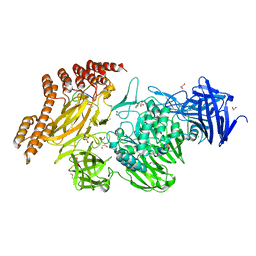 | | EngBF DARPin Fusion 4b G10 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Ernst, P, Pluckthun, A, Mittl, P.R.E. | | Deposit date: | 2019-01-10 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of biological targets by host:guest crystal lattice engineering.
Sci Rep, 9, 2019
|
|
4JO9
 
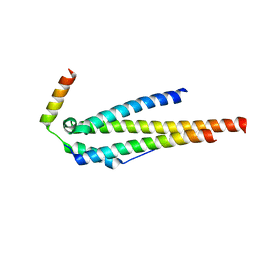 | | Crystal structure of the human Nup49CCS2+3* Nup57CCS3* complex 1:2 stoichiometry | | Descriptor: | Nucleoporin p54, Nucleoporin p58/p45 | | Authors: | Stuwe, T, Bley, C.J, Mayo, D.J, Hoelz, A. | | Deposit date: | 2013-03-18 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
1KZM
 
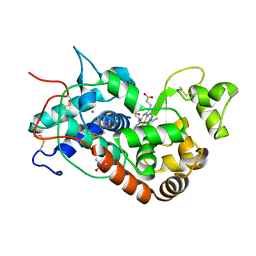 | | Distal Heme Pocket Mutant (R38S/H42E) of Recombinant Horseradish Peroxidase C (HRP C). | | Descriptor: | CACODYLATE ION, CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Meno, K, Henriksen, A, Smith, A.T, Gajhede, M. | | Deposit date: | 2002-02-07 | | Release date: | 2002-10-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of the two horseradish peroxidase catalytic residue variants H42E and R38S/H42E: implications for the catalytic cycle.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
6FLC
 
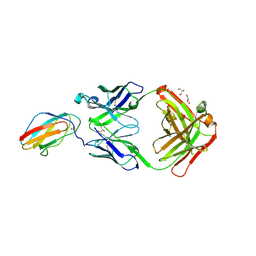 | | 2C8 Fab bound to EDIII of DenV 2 | | Descriptor: | Domain III of Dengue virus 2, GLYCEROL, Heavy chain of 2C8 Fab, ... | | Authors: | Flanagan, A, Renner, M, Grimes, J.M. | | Deposit date: | 2018-01-25 | | Release date: | 2018-10-24 | | Last modified: | 2018-10-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of a potent and highly unusual minimally enhancing antibody directed against dengue virus.
Nat. Immunol., 19, 2018
|
|
6QJD
 
 | |
7GSS
 
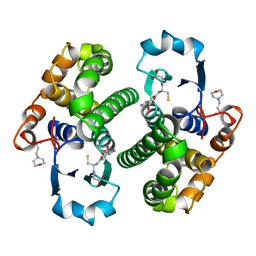 | | Human glutathione S-transferase P1-1, complex with glutathione | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, GLUTATHIONE S-TRANSFERASE P1-1 | | Authors: | Oakley, A, Parker, M. | | Deposit date: | 1997-08-13 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
3B2H
 
 | | Iodide derivative of human LFABP at high resolution | | Descriptor: | Fatty acid-binding protein, liver, IODIDE ION, ... | | Authors: | Sharma, A, Yogavel, M, Sharma, A. | | Deposit date: | 2011-08-03 | | Release date: | 2012-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Utility of anion and cation combinations for phasing of protein structures.
J.Struct.Funct.Genom., 13, 2012
|
|
6QJN
 
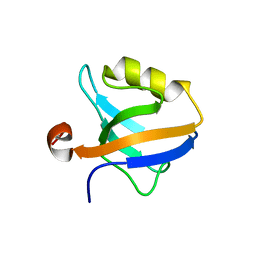 | |
6ZS3
 
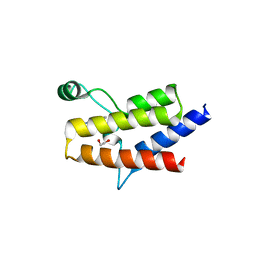 | | Crystal structure of the fifth bromodomain of human protein polybromo-1 in complex with 2-(6-amino-5-(piperazin-1-yl)pyridazin-3-yl)phenol | | Descriptor: | 1,2-ETHANEDIOL, 2-(6-azanyl-5-piperazin-4-ium-1-yl-pyridazin-3-yl)phenol, Protein polybromo-1 | | Authors: | Preuss, F, Joerger, A.C, Wanior, M, Kraemer, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-07-15 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Pan-SMARCA/PB1 Bromodomain Inhibitors and Their Role in Regulating Adipogenesis.
J.Med.Chem., 63, 2020
|
|
6FLA
 
 | | 3H5 Fab bound to EDIII of DenV 2 Xtal form 1 | | Descriptor: | CHLORIDE ION, Domain III of Dengue virus 2, GLYCEROL, ... | | Authors: | Flanagan, A, Renner, M, Grimes, J.M. | | Deposit date: | 2018-01-25 | | Release date: | 2018-10-24 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Characterization of a potent and highly unusual minimally enhancing antibody directed against dengue virus.
Nat. Immunol., 19, 2018
|
|
1I5V
 
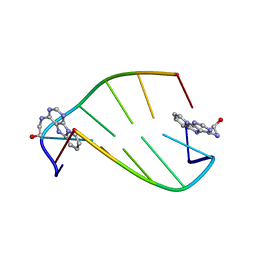 | | SOLUTION STRUCTURE OF 2-(PYRIDO[1,2-E]PURIN-4-YL)AMINO-ETHANOL INTERCALATED IN THE DNA DUPLEX D(CGATCG)2 | | Descriptor: | 2-(PYRIDO[1,2-E]PURIN-4-YL)AMINO-ETHANOL, 5'-D(*CP*GP*AP*TP*CP*G)-3' | | Authors: | Favier, A, Blackledge, M, Simorre, J.P, Marion, D, Debousy, J.C. | | Deposit date: | 2001-03-01 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of 2-(pyrido[1,2-e]purin-4-yl)amino-ethanol intercalated in the DNA duplex d(CGATCG)2.
Biochemistry, 40, 2001
|
|
