3OTR
 
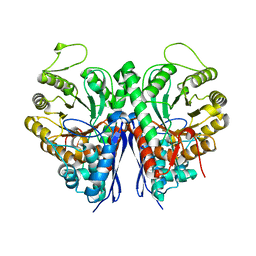 | | 2.75 Angstrom Crystal Structure of Enolase 1 from Toxoplasma gondii | | Descriptor: | CHLORIDE ION, Enolase, SULFATE ION | | Authors: | Minasov, G, Ruan, J, Shuvalova, L, Halavaty, A, Ngo, H, Tomavo, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-13 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structure of bradyzoite-specific enolase from Toxoplasma gondii reveals insights into its dual cytoplasmic and nuclear functions.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5UB8
 
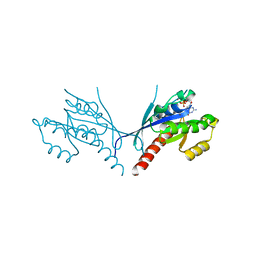 | | Crystal structure of YPT31, a Rab family GTPase from Candida albicans, in complex with GDP and Zn(II) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Likely rab family GTP-binding protein, ZINC ION | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-20 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of YPT31, a Rab family GTPase from Candida albicans, in complex with GDP and Zn(II)
To Be Published
|
|
5UE7
 
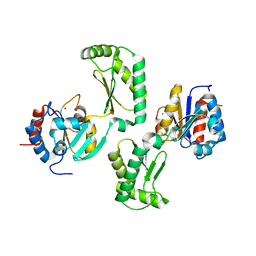 | | Crystal structure of the phosphomannomutase PMM1 from Candida albicans, apoenzyme state | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Phosphomannomutase | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-29 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the phosphomannomutase PMM1 from Candida albicans, apoenzyme state
To Be Published
|
|
5U08
 
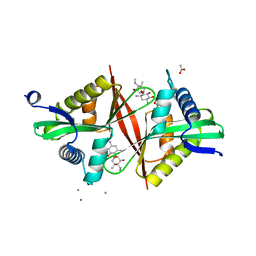 | | Crystal structure of an aminoglycoside acetyltransferase meta-AAC0020 from an uncultured soil metagenomic sample in complex with sisomicin | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, ACETATE ION, CALCIUM ION, ... | | Authors: | Xu, Z, Skarina, T, Wawrzak, Z, Stogios, P.J, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-23 | | Release date: | 2017-02-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural and Functional Survey of Environmental Aminoglycoside Acetyltransferases Reveals Functionality of Resistance Enzymes.
ACS Infect Dis, 3, 2017
|
|
3OUG
 
 | | Crystal structure of cleaved L-aspartate-alpha-decarboxylase from Francisella tularensis | | Descriptor: | Aspartate 1-decarboxylase, CHLORIDE ION, GLYCEROL | | Authors: | Nocek, B, Gu, M, Papazisi, L, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-14 | | Release date: | 2010-10-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of cleaved L-aspartate-alpha-decarboxylase from Francisella tularensis
To be Published
|
|
5UF8
 
 | | Crystal structure of the ARF family small GTPase ARF2 from Candida albicans in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Potential ADP-ribosylation factor | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-01-03 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.872 Å) | | Cite: | Crystal structure of the ARF family small GTPase ARF2 from Candida albicans in complex with GDP
To Be Published
|
|
3MSU
 
 | | Crystal Structure of Citrate Synthase from Francisella tularensis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Maltseva, N, Kim, Y, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-04-29 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.843 Å) | | Cite: | Crystal Structure of Citrate Synthase from Francisella tularensis
To be Published
|
|
5SWV
 
 | |
5UM7
 
 | | Crystal structure of the reduced state of the thiol-disulfide reductase SdbA from Streptococcus gordonii | | Descriptor: | ACETATE ION, Thioredoxin signature protein | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-01-26 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of the reduced state of the thiol-disulfide reductase SdbA from Streptococcus gordonii
To Be Published
|
|
3N77
 
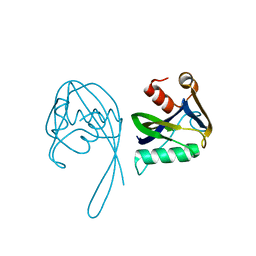 | | Crystal structure of Idp01880, putative NTP pyrophosphohydrolase of Salmonella typhimurium LT2 | | Descriptor: | Nucleoside triphosphatase nudI | | Authors: | Frydrysiak, J, Cooper, D.R, Derewenda, U, Anderson, W.F, Derewenda, Z.S, Integrated Center for Structure and Function Innovation (ISFI), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-26 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of Idp01880, putative NTP pyrophosphohydrolase of Salmonella typhimurium LT2
To be Published
|
|
3MZZ
 
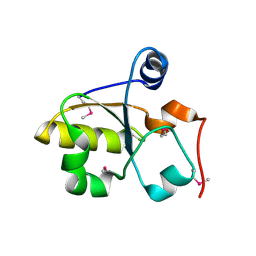 | | Crystal Structure of Rhodanese-like Domain Protein from Staphylococcus aureus | | Descriptor: | Rhodanese-like domain protein | | Authors: | Kim, Y, Chruszcz, M, Minor, W, Edwards, A, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-13 | | Release date: | 2010-06-30 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Rhodanese-like Domain Protein from Staphylococcus aureus
To be Published
|
|
3N0M
 
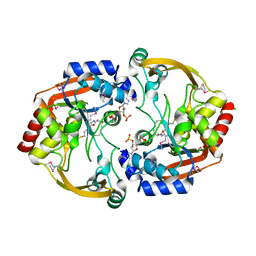 | | Crystal structure of BA2930 mutant (H183G) in complex with AcCoA | | Descriptor: | ACETYL COENZYME *A, Aminoglycoside N3-acetyltransferase, CHLORIDE ION | | Authors: | Klimecka, M.M, Chruszcz, M, Porebski, P.J, Cymborowski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-14 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Analysis of a Putative Aminoglycoside N-Acetyltransferase from Bacillus anthracis.
J.Mol.Biol., 410, 2011
|
|
3M84
 
 | | Crystal Structure of Phosphoribosylaminoimidazole Synthetase from Francisella tularensis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Maltseva, N, Kim, Y, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-17 | | Release date: | 2010-07-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal Structure of Phosphoribosylaminoimidazole Synthetase from
Francisella tularensis
To be Published
|
|
3P09
 
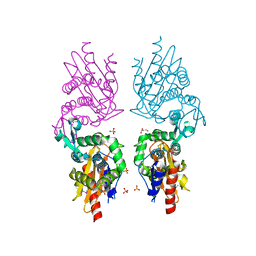 | | Crystal Structure of Beta-Lactamase from Francisella tularensis | | Descriptor: | Beta-lactamase, GLYCEROL, SULFATE ION | | Authors: | Kim, Y, Makowska-Grzyska, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-27 | | Release date: | 2010-10-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal Structure of Beta-Lactamase from Francisella tularensis
To be Published
|
|
3MXT
 
 | | Crystal Structure of Pantoate-Beta-alanine Ligase from Campylobacter jejuni | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Zhou, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-07 | | Release date: | 2010-06-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Pantoate-Beta-alanine Ligase from Campylobacter jejuni
To be Published
|
|
3N8H
 
 | | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis | | Descriptor: | ACETIC ACID, ADENOSINE MONOPHOSPHATE, GLYCEROL, ... | | Authors: | Maltseva, N, Kim, Y, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-28 | | Release date: | 2010-06-16 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis
To be Published, 2010
|
|
5UWY
 
 | | The crystal structure of thioredoxin reductase from Streptococcus pyogenes MGAS5005 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, Thioredoxin reductase | | Authors: | Tan, K, Zhou, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-21 | | Release date: | 2017-03-15 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | The crystal structure of thioredoxin reductase from Streptococcus pyogenes MGAS5005
To Be Published
|
|
5V0S
 
 | | Crystal structure of the ACT domain of prephenate dehydrogenase tyrA from Bacillus anthracis | | Descriptor: | CALCIUM ION, Prephenate dehydrogenase, SULFATE ION | | Authors: | Shabalin, I.G, Hou, J, Cymborowski, M.T, Otwinowski, Z, Kwon, K, Christendat, D, Gritsunov, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-28 | | Release date: | 2017-03-08 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and biochemical analysis of Bacillus anthracis prephenate dehydrogenase reveals an unusual mode of inhibition by tyrosine via the ACT domain.
Febs J., 287, 2020
|
|
3OS6
 
 | | Crystal structure of putative 2,3-dihydroxybenzoate-specific isochorismate synthase, DhbC from Bacillus anthracis. | | Descriptor: | GLYCEROL, Isochorismate synthase DhbC, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Domagalski, M.J, Chruszcz, M, Skarina, T, Onopriyenko, O, Cymborowski, M, Savchenko, A, Edwards, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-08 | | Release date: | 2010-10-20 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of isochorismate synthase DhbC from Bacillus anthracis.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3OSU
 
 | | Crystal structure of the 3-oxoacyl-acyl carrier protein reductase, FabG, from Staphylococcus aureus | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Edwards, A, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-09 | | Release date: | 2010-09-29 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the 3-oxoacyl-acyl carrier protein reductase, FabG, from Staphylococcus aureus
To be Published
|
|
3Q3V
 
 | | Crystal structure of Phosphoglycerate Kinase from Campylobacter jejuni. | | Descriptor: | FORMIC ACID, POTASSIUM ION, Phosphoglycerate kinase, ... | | Authors: | Filippova, E.V, Wawrzak, Z, Onopriyenko, O, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-22 | | Release date: | 2011-01-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.145 Å) | | Cite: | Crystal structures of putative phosphoglycerate kinases from B. anthracis and C. jejuni.
J.Struct.Funct.Genom., 13, 2012
|
|
5UCC
 
 | | Crystal structure of the ENTH domain of ENT2 from Candida albicans | | Descriptor: | CHLORIDE ION, CITRIC ACID, Potential epsin-like clathrin-binding protein | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-22 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of the ENTH domain of ENT2 from Candida albicans
To Be Published
|
|
3P4E
 
 | | Phosphoribosylformylglycinamidine cyclo-ligase from Vibrio cholerae | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, CITRIC ACID, ... | | Authors: | Osipiuk, J, Zhou, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-10-06 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Phosphoribosylformylglycinamidine cyclo-ligase from Vibrio cholerae.
To be Published
|
|
3OPK
 
 | | Crystal structure of divalent-cation tolerance protein CutA from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 | | Descriptor: | ACETATE ION, Divalent-cation tolerance protein cutA, MAGNESIUM ION, ... | | Authors: | Nocek, B, Mulligan, R, Papazisi, L, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-01 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of divalent-cation tolerance protein CutA from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2
TO BE PUBLISHED
|
|
3Q58
 
 | | Structure of N-acetylmannosamine-6-Phosphate Epimerase from Salmonella enterica | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-27 | | Release date: | 2011-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: |
|
|
