4MVE
 
 | |
4MZ8
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with an Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound C91 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-29 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5004 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound C91
To be Published
|
|
4MY1
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Bacillus anthracis str. Ames complexed with P68 | | Descriptor: | 1-(4-bromophenyl)-3-(2-{3-[(1E)-N-hydroxyethanimidoyl]phenyl}propan-2-yl)urea, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-26 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5997 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Bacillus anthracis str. Ames complexed with P68
To be Published
|
|
4N01
 
 | |
2A5Z
 
 | | Crystal Structure of Protein of Unknown Function SO2946 from Shewanella oneidensis MR-1 | | Descriptor: | MAGNESIUM ION, hypothetical protein SO2946 | | Authors: | Nocek, B, Bigelow, L, Abdullah, J, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-07-01 | | Release date: | 2005-08-16 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Structure of SO2946 orphan from Shewanella oneidensis shows "jelly-roll" fold with carbohydrate-binding module.
J.STRUCT.FUNCT.GENOM., 9, 2008
|
|
4J11
 
 | | The crystal structure of a secreted protein ESXB (wild-type, in P21 space group) from Bacillus anthracis str. sterne | | Descriptor: | SECRETED PROTEIN ESXB | | Authors: | Fan, Y, Tan, K, Chhor, G, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-31 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | The Crystal Structure of a Secreted Protein Esxb (Wild-Type, in P21 Space Group) from Bacillus Anthracis Str. Sterne
To be Published
|
|
3GE2
 
 | | Crystal structure of putative lipoprotein SP_0198 from Streptococcus pneumoniae | | Descriptor: | CHLORIDE ION, GLYCEROL, Lipoprotein, ... | | Authors: | Kim, Y, Zhang, R, Joachimiak, G, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-17 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Crystal Structure of Putative Lipoprotein SP_0198 from Streptococcus pneumoniae
To be Published
|
|
2AE8
 
 | | Crystal Structure of Imidazoleglycerol-phosphate Dehydratase from Staphylococcus aureus subsp. aureus N315 | | Descriptor: | Imidazoleglycerol-phosphate dehydratase, MAGNESIUM ION | | Authors: | Kim, Y, Quartey, P, Holzle, D, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-07-21 | | Release date: | 2005-09-06 | | Last modified: | 2015-05-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of Imidazoleglycerol-phosphate Dehydratase from Staphylococcus aureus subsp. aureus N315
To be Published
|
|
1Y5E
 
 | | Crystal structure of Molybdenum cofactor biosynthesis protein B | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, IMIDAZOLE, Molybdenum cofactor biosynthesis protein B | | Authors: | Chang, C, Zhou, M, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-02 | | Release date: | 2005-01-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Molybdenum cofactor biosynthesis protein B
TO BE PUBLISHED
|
|
4JBF
 
 | | Crystal structure of peptidoglycan glycosyltransferase from Atopobium parvulum DSM 20469. | | Descriptor: | Peptidoglycan glycosyltransferase, TETRAETHYLENE GLYCOL | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Shuvalova, L, Kiryukhina, O, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-20 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of peptidoglycan glycosyltransferase from Atopobium parvulum DSM 20469.
To be Published
|
|
1Y9B
 
 | | Structure of Conserved Putative Transcriptional Factor from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | conserved hypothetical protein | | Authors: | Binkowski, T.A, Hatzos, C, Quartey, P, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-15 | | Release date: | 2005-01-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conserverd hypothetical protein from Vibrio cholerae O1 biovar eltor str. N16961
To be Published
|
|
4NP6
 
 | | Crystal Structure of Adenylate Kinase from Vibrio cholerae O1 biovar eltor | | Descriptor: | Adenylate kinase | | Authors: | Kim, Y, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-20 | | Release date: | 2013-12-18 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Crystal Structure of Adenylate Kinase from Vibrio cholerae O1 biovar eltor
To be Published
|
|
1Y7U
 
 | | Crystal Structure of Acyl-Coa hydrolase from Bacillus cereus | | Descriptor: | Acyl-CoA hydrolase, CALCIUM ION, COENZYME A, ... | | Authors: | Kim, Y, Li, H, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-10 | | Release date: | 2005-01-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Acyl-CoA hydrolase from Bacillus cereus
To be Published
|
|
4NMY
 
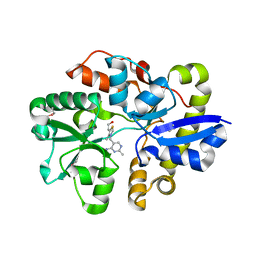 | | Crystal Structure of the Thiamin-bound form of Substrate-binding Protein of ABC Transporter from Clostridium difficile | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, ABC-type transport system, extracellular solute-binding protein | | Authors: | Kim, Y, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-15 | | Release date: | 2013-12-04 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Crystal Structure of the Thiamin-bound form of Substrate-binding Protein of ABC Transporter from Clostridium difficile
To be Published
|
|
4GPN
 
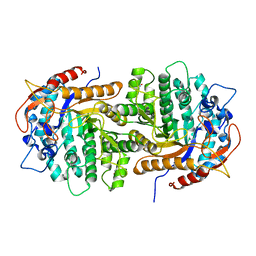 | | The crystal structure of 6-P-beta-D-Glucosidase (E375Q mutant) from Streptococcus mutans UA150 in complex with Gentiobiose 6-phosphate. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-O-phosphono-beta-D-glucopyranose-(1-6)-beta-D-glucopyranose, 6-phospho-beta-D-Glucosidase, ... | | Authors: | Tan, K, Michalska, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-08-21 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | GH1-family 6-P-beta-glucosidases from human microbiome lactic acid bacteria.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
1Z67
 
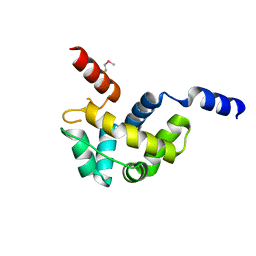 | | Structure of Homeodomain-like Protein of Unknown Function S4005 from Shigella flexneri | | Descriptor: | SODIUM ION, hypothetical protein S4005 | | Authors: | Osipiuk, J, Maltseva, N, Dementieva, I, Clancy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-03-21 | | Release date: | 2005-05-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of YidB protein from Shigella flexneri shows a new fold with homeodomain motif.
Proteins, 65, 2006
|
|
1Z72
 
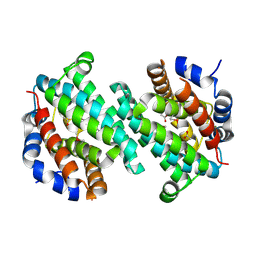 | | Structure of a putative transcriptional regulator from Streptococcus pneumoniae | | Descriptor: | ACETIC ACID, MAGNESIUM ION, transcriptional regulator, ... | | Authors: | Cuff, M.E, Zhou, M, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-03-23 | | Release date: | 2005-05-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a putative transcriptional regulator from Streptococcus pneumoniae
To be Published
|
|
4NZP
 
 | | The crystal structure of argininosuccinate synthase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | Argininosuccinate synthase | | Authors: | Tan, K, Gu, M, Zhang, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-12 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | The crystal structure of argininosuccinate synthase from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
4H0C
 
 | | Crystal structure of phospholipase/Carboxylesterase from Dyadobacter fermentans DSM 18053 | | Descriptor: | CITRIC ACID, GLYCEROL, Phospholipase/Carboxylesterase, ... | | Authors: | Chang, C, Holowicki, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-07 | | Release date: | 2012-09-26 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of phospholipase/Carboxylesterase from Dyadobacter fermentans DSM 18053
To be Published
|
|
4NCZ
 
 | | Spermidine N-acetyltransferase from Vibrio cholerae in complex with 2-[n-cyclohexylamino]ethane sulfonate. | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, SODIUM ION, ... | | Authors: | Osipiuk, J, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-10-25 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A Novel Polyamine Allosteric Site of SpeG from Vibrio cholerae Is Revealed by Its Dodecameric Structure.
J.Mol.Biol., 427, 2015
|
|
4NEG
 
 | | The crystal structure of tryptophan synthase subunit beta from Bacillus anthracis str. 'Ames Ancestor' | | Descriptor: | FORMIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Tan, K, Zhang, R, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-10-29 | | Release date: | 2013-11-13 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | The crystal structure of tryptophan synthase subunit beta from Bacillus anthracis str. 'Ames Ancestor'
To be Published
|
|
4HCI
 
 | | Uncharacterized Cupredoxin-like Domain Protein Cupredoxin_1 from Bacillus anthracis | | Descriptor: | Cupredoxin 1, GLYCEROL | | Authors: | Kim, Y, Maltseva, N, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-09-30 | | Release date: | 2012-10-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Uncharacterized Cupredoxin-like Domain Protein Cupredoxin_1 from Bacillus anthracis
To be Published
|
|
4MX8
 
 | |
2NS0
 
 | | Crystal structure of protein RHA04536 from Rhodococcus sp | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Hypothetical protein | | Authors: | Chang, C, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-11-02 | | Release date: | 2006-12-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Crystal structure of protein RHA04536 from Rhodococcus sp
To be Published
|
|
4JJT
 
 | | The crystal structure of enoyl-CoA hydratase from Mycobacterium tuberculosis H37Rv | | Descriptor: | ACETATE ION, Enoyl-CoA hydratase, GLYCEROL | | Authors: | Tan, K, Holowicki, J, Endres, M, Kim, C.-Y, Kim, H, Hung, L.-W, Terwilliger, T.C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2013-03-08 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.496 Å) | | Cite: | The crystal structure of enoyl-CoA hydratase from Mycobacterium tuberculosis H37Rv
To be Published
|
|
