2MNZ
 
 | | NMR Structure of KDM5B PHD1 finger in complex with H3K4me0(1-10aa) | | Descriptor: | H3K4me0, Lysine-specific demethylase 5B, ZINC ION | | Authors: | Zhang, Y, Yang, H.R, Guo, X, Rong, N.Y, Song, Y.J, Xu, Y.W, Lan, W.X, Xu, Y.H, Cao, C. | | Deposit date: | 2014-04-16 | | Release date: | 2014-08-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The PHD1 finger of KDM5B recognizes unmodified H3K4 during the demethylation of histone H3K4me2/3 by KDM5B.
Protein Cell, 5, 2014
|
|
8WD6
 
 | | Cryo-EM structure of the ABCG25 | | Descriptor: | ABC transporter G family member 25 | | Authors: | Xin, J, Yan, K.G. | | Deposit date: | 2023-09-14 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural insights into AtABCG25, an angiosperm-specific abscisic acid exporter.
Plant Commun., 5, 2024
|
|
8WBA
 
 | | Cryo-EM structure of the ABCG25 bound to CHS | | Descriptor: | ABC transporter G family member 25, CHOLESTEROL HEMISUCCINATE | | Authors: | Xin, J, Yan, K.G. | | Deposit date: | 2023-09-09 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into AtABCG25, an angiosperm-specific abscisic acid exporter.
Plant Commun., 5, 2024
|
|
8WAM
 
 | |
8WBX
 
 | | Cryo-EM structure of the ABCG25 bound to ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABC transporter G family member 25 | | Authors: | Xin, J, Yan, K.G. | | Deposit date: | 2023-09-10 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural insights into AtABCG25, an angiosperm-specific abscisic acid exporter.
Plant Commun., 5, 2024
|
|
1X84
 
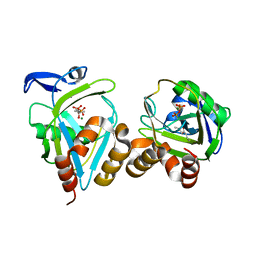 | | IPP isomerase (wt) reacted with (S)-bromohydrine of IPP | | Descriptor: | (S)-4-BROMO-3-HYDROXY-3-METHYLBUTYL DIPHOSPHATE, Isopentenyl-diphosphate delta-isomerase, MAGNESIUM ION, ... | | Authors: | Wouters, J, Oldfield, E. | | Deposit date: | 2004-08-17 | | Release date: | 2005-01-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A Crystallographic Investigation of Phosphoantigen Binding to Isopentenyl Pyrophosphate/Dimethylallyl Pyrophosphate Isomerase
J.Am.Chem.Soc., 127, 2005
|
|
1X83
 
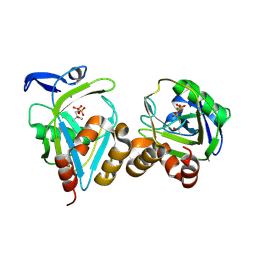 | | Y104F IPP isomerase reacted with (S)-bromohydrine of IPP | | Descriptor: | (S)-4-BROMO-3-HYDROXY-3-METHYLBUTYL DIPHOSPHATE, Isopentenyl-diphosphate delta-isomerase, MAGNESIUM ION, ... | | Authors: | Wouters, J, Oldfield, E. | | Deposit date: | 2004-08-17 | | Release date: | 2005-01-25 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Crystallographic Investigation of Phosphoantigen Binding to Isopentenyl Pyrophosphate/Dimethylallyl Pyrophosphate Isomerase
J.Am.Chem.Soc., 127, 2005
|
|
7BOR
 
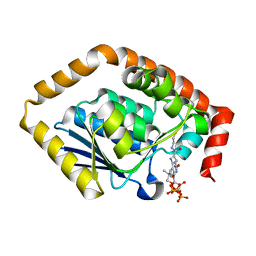 | | Structure of Pseudomonas aeruginosa CoA-bound OdaA | | Descriptor: | COENZYME A, Probable enoyl-CoA hydratase/isomerase | | Authors: | Zhao, N, Zhao, C, Liu, L, Li, T, Li, C, He, L, Zhu, Y, Song, Y, Bao, R. | | Deposit date: | 2020-03-19 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural and molecular dynamic studies of Pseudomonas aeruginosa OdaA reveal the regulation role of a C-terminal hinge element.
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
3EGT
 
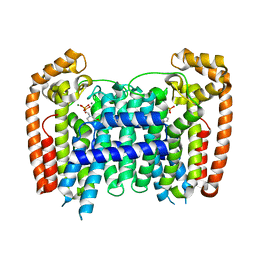 | | T. Brucei Farnesyl Diphosphate Synthase Complexed with Bisphosphonate BPH-722 | | Descriptor: | 1-(2,2-diphosphonoethyl)-3-(heptyloxy)pyridinium, Farnesyl pyrophosphate synthase, MAGNESIUM ION | | Authors: | Cao, R, Gao, Y, Robinson, H, Oldfield, E. | | Deposit date: | 2008-09-11 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lipophilic bisphosphonates as dual farnesyl/geranylgeranyl diphosphate synthase inhibitors: an X-ray and NMR investigation.
J.Am.Chem.Soc., 131, 2009
|
|
3EFQ
 
 | | T. Brucei Farnesyl Diphosphate Synthase Complexed with Bisphosphonate BPH-714 | | Descriptor: | 1-(2,2-diphosphonoethyl)-3-(octyloxy)pyridinium, Farnesyl pyrophosphate synthase, MAGNESIUM ION | | Authors: | Cao, R, Gao, Y, Robinson, H, Oldfield, E. | | Deposit date: | 2008-09-09 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lipophilic bisphosphonates as dual farnesyl/geranylgeranyl diphosphate synthase inhibitors: an X-ray and NMR investigation.
J.Am.Chem.Soc., 131, 2009
|
|
8H56
 
 | | Crystal structure of Rep' of porcine circovirus type 2 | | Descriptor: | Isoform Rep' of Replication-associated protein | | Authors: | Guan, S.Y, Song, Y.F. | | Deposit date: | 2022-10-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of the dimerized of porcine circovirus type II replication-related protein Rep'.
Proteins, 91, 2023
|
|
7BEM
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-269 scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-269 Vh domain, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEH
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-316 Fab | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-316 heavy chain, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEO
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in a ternary complex with COVOX-253H55L and COVOX-75 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEN
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in a ternary complex with COVOX-253 and COVOX-75 Fabs | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEI
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-150 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-150 heavy chain, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEP
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in a ternary complex with COVOX-384 and S309 Fabs | | Descriptor: | CHLORIDE ION, COVOX-384 heavy chain, COVOX-384 light chain, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEJ
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-158 Fab (crystal form 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-158 heavy chain, COVOX-158 light chain, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEL
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in a ternary complex with COVOX-88 and COVOX-45 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, COVOX-45 heavy chain, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEK
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-158 Fab (crystal form 2) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
3DYH
 
 | | T. Brucei Farnesyl Diphosphate Synthase Complexed with Bisphosphonate BPH-721 | | Descriptor: | 3-butoxy-1-(2,2-diphosphonoethyl)pyridinium, Farnesyl pyrophosphate synthase, MAGNESIUM ION | | Authors: | Cao, R, Gao, Y, Robinson, H, Goddard, A, Oldfield, E. | | Deposit date: | 2008-07-27 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Lipophilic bisphosphonates as dual farnesyl/geranylgeranyl diphosphate synthase inhibitors: an X-ray and NMR investigation.
J.Am.Chem.Soc., 131, 2009
|
|
3DYF
 
 | | T. Brucei Farnesyl Diphosphate Synthase Complexed with Bisphosphonate BPH-461 and Isopentyl Diphosphate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-FLUORO-1-(2-HYDROXY-2,2-DIPHOSPHONOETHYL)PYRIDINIUM, ACETATE ION, ... | | Authors: | Cao, R, Gao, Y, Robinson, H, Goddard, A, Oldfield, E. | | Deposit date: | 2008-07-27 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Lipophilic bisphosphonates as dual farnesyl/geranylgeranyl diphosphate synthase inhibitors: an X-ray and NMR investigation.
J.Am.Chem.Soc., 131, 2009
|
|
3DYG
 
 | | T. Brucei Farnesyl Diphosphate Synthase Complexed with Bisphosphonate BPH-461 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-FLUORO-1-(2-HYDROXY-2,2-DIPHOSPHONOETHYL)PYRIDINIUM, ACETATE ION, ... | | Authors: | Cao, R, Gao, Y, Robinson, H, Goddard, A, Oldfield, E. | | Deposit date: | 2008-07-27 | | Release date: | 2009-05-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Lipophilic bisphosphonates as dual farnesyl/geranylgeranyl diphosphate synthase inhibitors: an X-ray and NMR investigation.
J.Am.Chem.Soc., 131, 2009
|
|
7LUK
 
 | | CRYSTAL STRUCTURE OF RAR-RELATED ORPHAN RECEPTOR C (NHIS-RORGT(244-487)-L6-SRC1(678-692) IN COMPLEX WITH AN AZATRICYCLIC RORGT INVERSE AGONIST | | Descriptor: | (2S)-N-[(6aS,7R,9aS)-9a-[(4-fluorophenyl)sulfonyl]-3-(1,1,1,2,3,3,3-heptafluoropropan-2-yl)-6,6a,7,8,9,9a-hexahydro-5H-cyclopenta[f]quinolin-7-yl]-2-hydroxy-2-methyl-3-(methylsulfonyl)propanamide, Nuclear receptor ROR-gamma | | Authors: | Sack, J.S. | | Deposit date: | 2021-02-22 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.087 Å) | | Cite: | Azatricyclic Inverse Agonists of ROR gamma t That Demonstrate Efficacy in Models of Rheumatoid Arthritis and Psoriasis.
Acs Med.Chem.Lett., 12, 2021
|
|
7QHE
 
 | | Human Butyrylcholinesterase in complex with (S)-1-(4-((naphthalen-1-yl)carbamoyl)benzyl)-N-(3-((1,2,3,4-tetrahydroacridin-9-yl)amino)propyl)piperidine-3-carboxamide | | Descriptor: | (3~{S})-1-[[4-(naphthalen-1-ylcarbamoyl)phenyl]methyl]-~{N}-[3-(1,2,3,4-tetrahydroacridin-9-ylamino)propyl]piperidine-3-carboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Jing, L, Zhan, P, Liu, X, Nachon, F. | | Deposit date: | 2021-12-12 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Rapid discovery and crystallography study of highly potent and selective butylcholinesterase inhibitors based on oxime-containing libraries and conformational restriction strategies.
Bioorg.Chem., 134, 2023
|
|
