8B0L
 
 | | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with PE | | Descriptor: | Apolipoprotein N-acyltransferase, PHOSPHATIDYLETHANOLAMINE | | Authors: | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8AQ3
 
 | | In surfo structure of the membrane integral lipoprotein N-acyltransferase Lnt from E. coli in complex with PE | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Huang, C.-Y, Weichert, D, Boland, C, Smithers, L, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2022-08-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8B0K
 
 | | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli (Apo form) | | Descriptor: | Apolipoprotein N-acyltransferase | | Authors: | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8AQ2
 
 | | In meso structure of the membrane integral lipoprotein N-acyltransferase Lnt from P. aeruginosa covalently linked with TITC | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apolipoprotein N-acyltransferase, CITRATE ANION, ... | | Authors: | Huang, C.-Y, Weichert, D, Boland, C, Smithers, L, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2022-08-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8B0N
 
 | | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with Lyso-PE | | Descriptor: | Apolipoprotein N-acyltransferase, [(2~{S})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-oxidanyl-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8B0P
 
 | | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with Pam3 | | Descriptor: | Apolipoprotein N-acyltransferase, Pam3-SKKKK, [(2~{S})-3-[(2~{S})-3-azanyl-2-(hexadecanoylamino)-3-oxidanylidene-propyl]sulfanyl-2-hexadecanoyloxy-propyl] hexadecanoate | | Authors: | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8B0M
 
 | | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with PE (C387S mutant) | | Descriptor: | Apolipoprotein N-acyltransferase, PHOSPHATIDYLETHANOLAMINE | | Authors: | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8B0O
 
 | | Cryo-EM structure apolipoprotein N-acyltransferase Lnt from E.coli in complex with FP3 | | Descriptor: | Apolipoprotein N-acyltransferase, [(2~{R})-3-[(2~{R})-3-[[(2~{R})-1-[[(2~{R})-1-[[(2~{R})-6-[(2-aminophenyl)carbonylamino]-1-azanyl-1-oxidanylidene-hexan-2-yl]amino]-3-oxidanyl-1-oxidanylidene-propan-2-yl]amino]-3-oxidanyl-1-oxidanylidene-propan-2-yl]amino]-2-(hexadecanoylamino)-3-oxidanylidene-propyl]sulfanyl-2-hexadecanoyloxy-propyl] hexadecanoate | | Authors: | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8AQ4
 
 | | In surfo structure of the membrane integral lipoprotein N-acyltransferase Lnt from E. coli in complex with TITC and lyso-PE | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Huang, C.-Y, Weichert, D, Boland, C, Smithers, L, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2022-08-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
6O9U
 
 | | KirBac3.1 at a resolution of 2 Angstroms | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3,3',3''-phosphoryltripropanoic acid, BARIUM ION, ... | | Authors: | Gulbis, J.M, Clarke, O.B. | | Deposit date: | 2019-03-15 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A constricted opening in Kir channels does not impede potassium conduction.
Nat Commun, 11, 2020
|
|
6OR2
 
 | | MmpL3 is a lipid transporter that binds trehalose monomycolate and phosphatidylethanolamine | | Descriptor: | (1S)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, DODECYL-BETA-D-MALTOSIDE, Membrane protein, ... | | Authors: | Su, C.-C. | | Deposit date: | 2019-04-29 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | MmpL3 is a lipid transporter that binds trehalose monomycolate and phosphatidylethanolamine.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OQV
 
 | | E. coli ATP Synthase State 2b | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2019-04-29 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
6Y5V
 
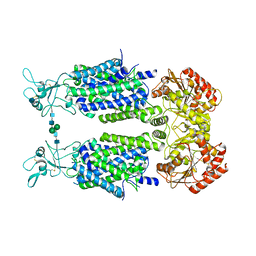 | | Structure of Human Potassium Chloride Transporter KCC3b (S45D/T940D/T997D) in KCl | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier family 12 member 6, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chi, G, Man, H, Ebenhoch, R, Reggiano, G, Pike, A.C.W, Wang, D, McKinley, G, Mukhopadhyay, S.M.M, MacLean, E.M, Chalk, R, Moreau, C, Snee, M, Bohstedt, T, Singh, N.K, Abrusci, P, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Marsden, B.D, Burgess-Brown, N.A, DiMaio, F, Duerr, K.L. | | Deposit date: | 2020-02-25 | | Release date: | 2020-07-15 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Phospho-regulation, nucleotide binding and ion access control in potassium-chloride cotransporters.
Embo J., 40, 2021
|
|
6Y0P
 
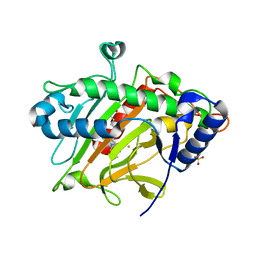 | | isopenicillin N synthase in complex with IPN and Fe using FT-SSX methods | | Descriptor: | FE (III) ION, ISOPENICILLIN N, Isopenicillin N synthase, ... | | Authors: | Rabe, P, Beale, J.H, Lang, P.A, Dirr, A.S, Leissing, T.M, Butryn, A, Aller, P, Kamps, J.J.A.G, Axford, D, McDonough, M.A, Orville, A.M, Owen, R, Schofield, C.J. | | Deposit date: | 2020-02-10 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-ray free-electron laser studies reveal correlated motion during isopenicillin N synthase catalysis.
Sci Adv, 7, 2021
|
|
6Z3Y
 
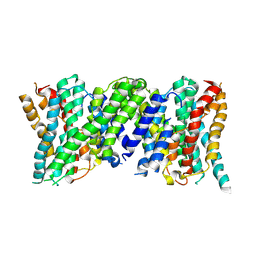 | |
6Z7P
 
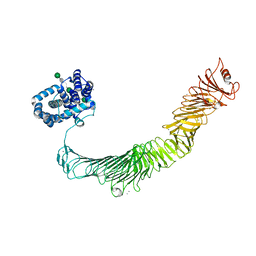 | | Composite model of the Caulobacter crescentus S-layer bound to the O-antigen of lipopolysaccharide | | Descriptor: | 4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose, CALCIUM ION, S-layer protein | | Authors: | Bharat, T.A.M, von Kugelgen, A. | | Deposit date: | 2020-06-01 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | In Situ Structure of an Intact Lipopolysaccharide-Bound Bacterial Surface Layer.
Cell, 180, 2020
|
|
6Z9A
 
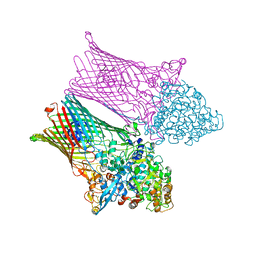 | | Fructo-oligosaccharide transporter BT 1762-63 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, SusC homolog, ... | | Authors: | van den Berg, B. | | Deposit date: | 2020-06-03 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Insights into SusCD-mediated glycan import by a prominent gut symbiont.
Nat Commun, 12, 2021
|
|
6ZAZ
 
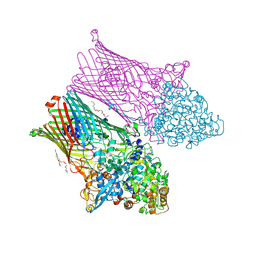 | | Fructo-oligosaccharide transporter BT 1762-63 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | van den Berg, B. | | Deposit date: | 2020-06-06 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Insights into SusCD-mediated glycan import by a prominent gut symbiont.
Nat Commun, 12, 2021
|
|
6Z8I
 
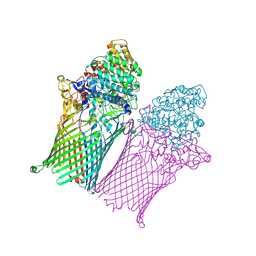 | | Fructo-oligosaccharide transporter BT 1762-63 | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, SusC homolog, ... | | Authors: | van den Berg, B. | | Deposit date: | 2020-06-02 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Insights into SusCD-mediated glycan import by a prominent gut symbiont.
Nat Commun, 12, 2021
|
|
6Z3Z
 
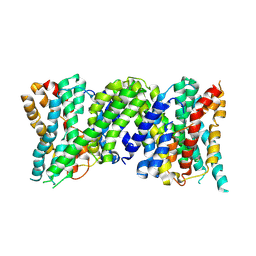 | |
5NDA
 
 | | NMR Structural Characterisation of Pharmaceutically Relevant Proteins Obtained Through a Novel Recombinant Production: The Case of The Pulmonary Surfactant Polypeptide C Analogue rSP-C33Leu. | | Descriptor: | rSP-C33Leu -RECOMBINANT PULMONARY SURFACTANT-ASSOCIATED POLYPEPTIDE C ANALOGUE- | | Authors: | Venturi, L, Pioselli, B, Johansson, J, Rising, A, Kronqvist, N, Nordling, K. | | Deposit date: | 2017-03-08 | | Release date: | 2017-06-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Efficient protein production inspired by how spiders make silk.
Nat Commun, 8, 2017
|
|
5O65
 
 | | Crystal Structure of the Pseudomonas functional amyloid secretion protein FapF | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, FapF, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Rouse, S.L, Hare, S, Lambert, S, Morgan, R.M.L, Hawthorne, W.J, Berry, J, Matthews, S.J. | | Deposit date: | 2017-06-05 | | Release date: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A new class of hybrid secretion system is employed in Pseudomonas amyloid biogenesis.
Nat Commun, 8, 2017
|
|
7E33
 
 | | Serotonin 1E (5-HT1E) receptor-Gi protein complex | | Descriptor: | 3-(1-methylpiperidin-4-yl)-1H-indol-5-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-14 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
7E2Y
 
 | | Serotonin-bound Serotonin 1A (5-HT1A) receptor-Gi protein complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-14 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
7E2Z
 
 | | Aripiprazole-bound serotonin 1A (5-HT1A) receptor-Gi protein complex | | Descriptor: | 7-[4-[4-[2,3-bis(chloranyl)phenyl]piperazin-1-yl]butoxy]-3,4-dihydro-1H-quinolin-2-one, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-14 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
