7MQ5
 
 | | Crystal Structure of Putative Universal Stress Protein from Pseudomonas aeruginosa UCBPP-PA14 | | Descriptor: | CHLORIDE ION, Universal stress protein | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of Putative Universal Stress Protein from Pseudomonas aeruginosa UCBPP-PA14
To Be Published
|
|
4JG2
 
 | | Structure of phage-related protein from Bacillus cereus ATCC 10987 | | Descriptor: | Phage-related protein | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Shuvalova, L, Kiryukhina, O, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2013-02-28 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of phage-related protein from Bacillus cereus ATCC 10987
To be Published
|
|
7KPO
 
 | | High Resolution Crystal Structure of the DNA-binding Domain from the Sensor Histidine Kinase ChiS from Vibrio cholerae | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, Response regulator, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-12 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | High Resolution Crystal Structure of the DNA-binding Domain from the Sensor Histidine Kinase ChiS from Vibrio cholerae.
To Be Published
|
|
4KMR
 
 | | Structure of a putative transcriptional regulator of LacI family from Sanguibacter keddieii DSM 10542. | | Descriptor: | MAGNESIUM ION, SODIUM ION, Transcriptional regulator, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-08 | | Release date: | 2013-06-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a putative transcriptional regulator of LacI family from Sanguibacter keddieii DSM 10542.
To be Published
|
|
7KOM
 
 | | High Resolution Crystal Structure of Putative Pterin Binding Protein PruR (VV2_1280) from Vibrio vulnificus CMCP6 | | Descriptor: | FORMIC ACID, Oxidored_molyb domain-containing protein, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-09 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | High Resolution Crystal Structure of Putative Pterin Binding Protein PruR (VV2_1280) from Vibrio vulnificus CMCP6.
To Be Published
|
|
7KP2
 
 | | High Resolution Crystal Structure of Putative Pterin Binding Protein (PruR) from Vibrio cholerae O1 biovar El Tor str. N16961 in Complex with Neopterin | | Descriptor: | L-NEOPTERIN, Putative Pterin Binding Protein | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Pshenychnyi, S, Dubrovska, I, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-10 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | High Resolution Crystal Structure of Putative Pterin Binding Protein (PruR) from Vibrio cholerae O1 biovar El Tor str. N16961 in Complex with Neopterin.
To Be Published
|
|
4ISC
 
 | | Crystal structure of a putative Methyltransferase from Pseudomonas syringae | | Descriptor: | BETA-MERCAPTOETHANOL, Methyltransferase | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-16 | | Release date: | 2013-02-20 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of a putative Methyltransferase from Pseudomonas syringae
To be Published
|
|
4KWA
 
 | | Crystal structure of a putative transcriptional regulator from Saccharomonospora viridis in complex with choline | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, CHOLINE ION, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-23 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a putative transcriptional regulator from Saccharomonospora viridis in complex with choline
To be Published
|
|
4JBE
 
 | | 1.95 Angstrom Crystal Structure of Gamma-glutamyl phosphate Reductase from Saccharomonospora viridis. | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-MERCAPTOETHANOL, ... | | Authors: | Minasov, G, Filippova, E.V, Halavaty, A, Shuvalova, L, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Crystal Structure of Gamma-glutamyl phosphate Reductase from Saccharomonospora viridis.
TO BE PUBLISHED
|
|
4R87
 
 | | Crystal structure of spermidine N-acetyltransferase from Vibrio cholerae in complex with CoA and spermine | | Descriptor: | COENZYME A, DI(HYDROXYETHYL)ETHER, SPERMINE, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Kuhn, M.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-29 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | A Novel Polyamine Allosteric Site of SpeG from Vibrio cholerae Is Revealed by Its Dodecameric Structure.
J.Mol.Biol., 427, 2015
|
|
4HVN
 
 | | Crystal structure of hypothetical protein with ketosteroid isomerase-like protein fold from Catenulispora acidiphila DSM 44928 in complex with Trimethylamine. | | Descriptor: | N,N-dimethylmethanamine, hypothetical protein | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-06 | | Release date: | 2012-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of a hypothetical protein: a potential agent involved in trimethylamine metabolism in Catenulispora acidiphila.
J.Struct.Funct.Genom., 15, 2014
|
|
6BQ9
 
 | | 2.55 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-493) of DNA Topoisomerase IV Subunit A from Pseudomonas putida | | Descriptor: | CHLORIDE ION, DNA topoisomerase 4 subunit A, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-27 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | 2.55 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-493) of DNA Topoisomerase IV Subunit A from Pseudomonas putida.
To Be Published
|
|
6C4V
 
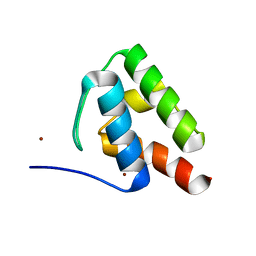 | | 1.9 Angstrom Resolution Crystal Structure of Acyl Carrier Protein Domain (residues 1350-1461) of Polyketide Synthase Pks13 from Mycobacterium tuberculosis | | Descriptor: | Polyketide synthase Pks13, ZINC ION | | Authors: | Minasov, G, Brunzelle, J.S, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-01-12 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Resolution Crystal Structure of Acyl Carrier Protein Domain (residues 1350-1461) of Polyketide Synthase Pks13 from Mycobacterium tuberculosis.
To be Published
|
|
6CY6
 
 | | Crystal structure of spermidine/spermine N-acetyltransferase SpeG from Escherichia coli in complex with tris(hydroxymethyl)aminomethane. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-04 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Analysis of crystalline and solution states of ligand-free spermidine N-acetyltransferase (SpeG) from Escherichia coli.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6D0P
 
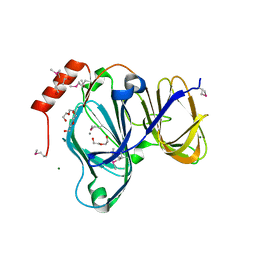 | | 1.88 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Dubrovska, I, Kiryukhina, O, Endres, M, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-10 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 1.88 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii
To be Published
|
|
6D0G
 
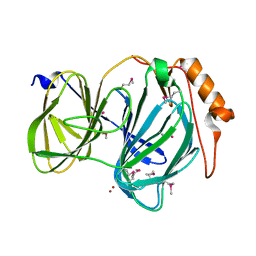 | | 1.78 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii | | Descriptor: | BROMIDE ION, MANGANESE (II) ION, Pirin family protein | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Dubrovska, I, Kiryukhina, O, Endres, M, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-10 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | 1.78 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii.
To be Published
|
|
6DB1
 
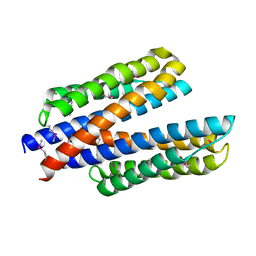 | | 2.0 Angstrom Resolution Crystal Structure of N-Terminal Ligand-Binding Domain of Putative Methyl-Accepting Chemotaxis Protein from Salmonella enterica | | Descriptor: | CHLORIDE ION, Putative methyl-accepting chemotaxis protein | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 Angstrom Resolution Crystal Structure of N-Terminal Ligand-Binding Domain of Putative Methyl-Accepting Chemotaxis Protein from Salmonella enterica.
To Be Published
|
|
6D72
 
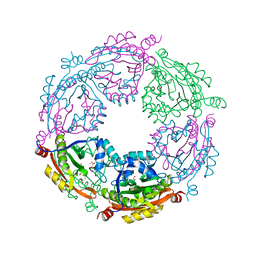 | | Crystal structure of spermidine/spermine N-acetyltransferase SpeG from Yersinia pestis in complex with calcium ions. | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, MALONATE ION, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-23 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of spermidine/spermine N-acetyltransferase SpeG from Yersinia pestis in complex with calcium ions.
To Be Published
|
|
6C4Q
 
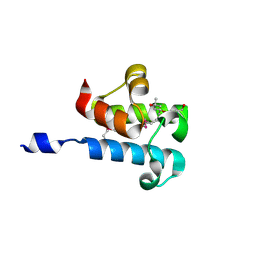 | | 1.16 Angstrom Resolution Crystal Structure of Acyl Carrier Protein Domain (residues 1-100) of Polyketide Synthase Pks13 from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Polyketide synthase Pks13 | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-01-12 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | 1.16 Angstrom Resolution Crystal Structure of Acyl Carrier Protein Domain (residues 1-100) of Polyketide Synthase Pks13 from Mycobacterium tuberculosis.
To Be Published
|
|
6CMZ
 
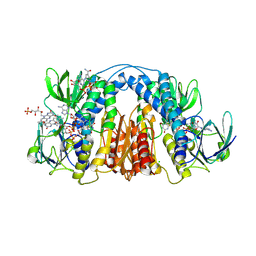 | | 2.3 Angstrom Resolution Crystal Structure of Dihydrolipoamide Dehydrogenase from Burkholderia cenocepacia in Complex with FAD and NAD | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, D-MALATE, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-03-06 | | Release date: | 2018-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2.3 Angstrom Resolution Crystal Structure of Dihydrolipoamide Dehydrogenase from Burkholderia cenocepacia in Complex with FAD and NAD.
To Be Published
|
|
6BXG
 
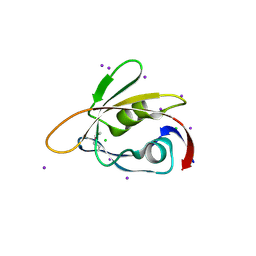 | | 1.45 Angstrom Resolution Crystal Structure of PDZ domain of Carboxy-Terminal Protease from Vibrio cholerae in Complex with Peptide. | | Descriptor: | CHLORIDE ION, IODIDE ION, LEU-ILE-ALA, ... | | Authors: | Minasov, G, Shuvalova, L, Filippova, E.V, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-12-18 | | Release date: | 2018-01-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | 1.45 Angstrom Resolution Crystal Structure of PDZ domain of Carboxy-Terminal Protease from Vibrio cholerae in Complex with Peptide.
To Be Published
|
|
6DVV
 
 | | 2.25 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Klebsiella pneumoniae in Complex with NAD and Mn2+. | | Descriptor: | 6-phospho-alpha-glucosidase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-25 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6DXN
 
 | | 1.95 Angstrom Resolution Crystal Structure of DsbA Disulfide Interchange Protein from Klebsiella pneumoniae. | | Descriptor: | TRIETHYLENE GLYCOL, Thiol:disulfide interchange protein | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Kiryukhina, O, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-29 | | Release date: | 2018-07-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6DLL
 
 | | 2.2 Angstrom Resolution Crystal Structure of P-Hydroxybenzoate Hydroxylase from Pseudomonas putida in Complex with FAD. | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-01 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural comparison of p-hydroxybenzoate hydroxylase (PobA) from Pseudomonas putida with PobA from other Pseudomonas spp. and other monooxygenases.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6BZ0
 
 | | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Acinetobacter baumannii in Complex with FAD. | | Descriptor: | CHLORIDE ION, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-12-21 | | Release date: | 2018-01-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Acinetobacter baumannii in Complex with FAD.
To Be Published
|
|
