2X6Y
 
 | | Tailspike protein mutant D339A of E.coli bacteriophage HK620 in complex with hexasaccharide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SODIUM ION, TAIL SPIKE PROTEIN, ... | | Authors: | Lorenzen, N.K, Mueller, J.J, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2010-02-22 | | Release date: | 2011-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Single Amino Acid Exchange in Bacteriophage Hk620 Tailspike Protein Results in Thousand-Fold Increase of its Oligosaccharide Affinity.
Glycobiology, 23, 2013
|
|
2X85
 
 | | Tailspike protein of E. coli bacteriophage HK620 in complex with hexasaccharide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SODIUM ION, TAILSPIKE PROTEIN HK620, ... | | Authors: | Lorenzen, N.K, Mueller, J.J, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2010-03-05 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Single Amino Acid Exchange in Bacteriophage Hk620 Tailspike Protein Results in Thousand-Fold Increase of its Oligosaccharide Affinity.
Glycobiology, 23, 2013
|
|
3PF4
 
 | | Crystal structure of Bs-CspB in complex with r(GUCUUUA) | | Descriptor: | Cold shock protein cspB, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Sachs, R, Max, K.E.A, Heinemann, U. | | Deposit date: | 2010-10-27 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | RNA single strands bind to a conserved surface of the major cold shock protein in crystals and solution.
Rna, 18, 2012
|
|
3PF5
 
 | |
3PDO
 
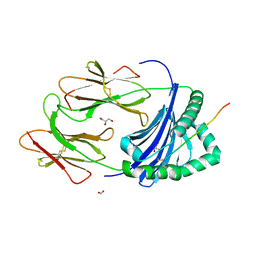 | | Crystal Structure of HLA-DR1 with CLIP102-120 | | Descriptor: | FORMIC ACID, GLYCEROL, HLA class II histocompatibility antigen gamma chain, ... | | Authors: | Gunther, S, Schlundt, A, Sticht, J, Roske, Y, Heinemann, U, Wiesmuller, K.-H, Jung, G, Falk, K, Rotzschke, O, Freund, C. | | Deposit date: | 2010-10-23 | | Release date: | 2010-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Bidirectional binding of invariant chain peptides to an MHC class II molecule.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3PGC
 
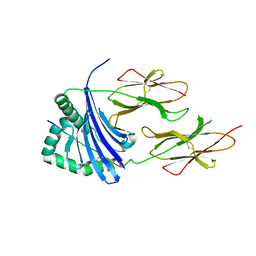 | | Crystal Structure of HLA-DR1 with CLIP106-120, flipped peptide orientation | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, HLA class II histocompatibility antigen gamma chain, ... | | Authors: | Gunther, S, Schlundt, A, Sticht, J, Roske, Y, Heinemann, U, Wiesmuller, K.-H, Jung, G, Falk, K, Rotzschke, O, Freund, C. | | Deposit date: | 2010-11-01 | | Release date: | 2010-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Bidirectional binding of invariant chain peptides to an MHC class II molecule.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3PGD
 
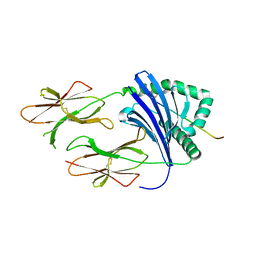 | | Crystal Structure of HLA-DR1 with CLIP106-120, canonical peptide orientation | | Descriptor: | HLA class II histocompatibility antigen gamma chain, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Gunther, S, Schlundt, A, Sticht, J, Roske, Y, Heinemann, U, Wiesmuller, K.-H, Jung, G, Falk, K, Rotzschke, O, Freund, C. | | Deposit date: | 2010-11-01 | | Release date: | 2010-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Bidirectional binding of invariant chain peptides to an MHC class II molecule.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1KQ6
 
 | |
1XCR
 
 | | Crystal Structure of Longer Splice Variant of PTD012 from Homo sapiens reveals a novel Zinc-containing fold | | Descriptor: | ACETIC ACID, ZINC ION, hypothetical protein PTD012 | | Authors: | Manjasetty, B.A, Fieber-Erdmann, M, Roske, Y, Goetz, F, Buessow, K, Heinemann, U. | | Deposit date: | 2004-09-03 | | Release date: | 2005-09-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Homo sapiens PTD012 reveals a zinc-containing hydrolase fold
Protein Sci., 15, 2006
|
|
6H0Z
 
 | | Crystal Structure of KDM4D with tetrazolylhydrazide ligand NR067 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Weiss, M.S, Heinemann, U, Link, A. | | Deposit date: | 2018-07-10 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal Structure of KDM4D with tetrazolylhydrazide ligand NR067
To be published
|
|
6HQC
 
 | |
6ZC6
 
 | |
6ZBQ
 
 | | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling | | Descriptor: | 5-fluoranyl-2-(5,6,7,8-tetrahydronaphthalen-2-ylsulfonylamino)benzoic acid, Dishevelled, dsh homolog 3 (Drosophila), ... | | Authors: | Roske, Y, Heinemann, U, Oschkinat, H. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling
J.Magn.Reson., 2021
|
|
6ZBZ
 
 | | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3,4-dimethylphenyl)sulfonylamino]-5-fluoranyl-benzoic acid, Dishevelled, ... | | Authors: | Roske, Y, Heinemann, U, Oschkinat, H. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling
J.Magn.Reson., 2021
|
|
6ZC7
 
 | |
6ZC3
 
 | | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling | | Descriptor: | 1,2-ETHANEDIOL, 5-fluoranyl-2-(methylsulfonylamino)benzoic acid, Dishevelled, ... | | Authors: | Roske, Y, Heinemann, U, Oschkinat, H. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling
J.Magn.Reson., 2021
|
|
6ZC8
 
 | |
6ZC4
 
 | | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling | | Descriptor: | 5-bromanyl-2-(5,6,7,8-tetrahydronaphthalen-2-ylsulfonylamino)benzoic acid, Dishevelled, dsh homolog 3 (Drosophila), ... | | Authors: | Roske, Y, Heinemann, U, Oschkinat, H. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling
J.Magn.Reson., 2021
|
|
6H0X
 
 | | Crystal Structure of KDM4D with tetrazolylhydrazide ligand AA040 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Weiss, M.S, Heinemann, U, Link, A. | | Deposit date: | 2018-07-10 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structure of KDM4D with tetrazolylhydrazide ligand AA040
To be published
|
|
6H11
 
 | | Crystal Structure of KDM4D with tetrazolylhydrazide ligand AA028 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Weiss, M.S, Heinemann, U, Link, A. | | Deposit date: | 2018-07-10 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.516 Å) | | Cite: | Crystal Structure of KDM4D with tetrazolylhydrazide ligand AA028
To be published
|
|
6H0W
 
 | | Crystal Structure of KDM4D with tetrazolylhydrazide ligand NS035 | | Descriptor: | (2~{R})-3-phenyl-2-(2~{H}-1,2,3,4-tetrazol-5-yl)propanehydrazide, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Malecki, P.H, Weiss, M.S, Heinemann, U, Link, A. | | Deposit date: | 2018-07-10 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Crystal Structure of KDM4D with tetrazolylhydrazide ligand NS035
To be published
|
|
6H0Y
 
 | | Crystal Structure of KDM4D with tetrazolylhydrazide ligand NS022 | | Descriptor: | (2~{R})-3-(4-methoxyphenyl)-2-(2~{H}-1,2,3,4-tetrazol-5-yl)propanehydrazide, 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Weiss, M.S, Heinemann, U, Link, A. | | Deposit date: | 2018-07-10 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.212 Å) | | Cite: | Crystal Structure of KDM4D with tetrazolylhydrazide ligand NS022
To be published
|
|
6H10
 
 | | Crystal Structure of KDM4D with tetrazolylhydrazide ligand NR073 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Weiss, M.S, Heinemann, U, Link, A. | | Deposit date: | 2018-07-10 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.104 Å) | | Cite: | Crystal Structure of KDM4D with tetrazolylhydrazide ligand NR073
To be published
|
|
466D
 
 | | DISORDER AND TWIN REFINEMENT OF RNA HEPTAMER DOUBLE HELIX | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*CP*UP*A)-3'), RNA (5'-R(*UP*AP*GP*CP*UP*CP*C)-3'), SODIUM ION, ... | | Authors: | Mueller, U, Muller, Y.A, Herbst-Irmer, R, Sprinzl, M, Heinemann, U. | | Deposit date: | 1999-04-14 | | Release date: | 1999-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Disorder and twin refinement of RNA heptamer double helices.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1I5F
 
 | | BACILLUS CALDOLYTICUS COLD-SHOCK PROTEIN MUTANTS TO STUDY DETERMINANTS OF PROTEIN STABILITY | | Descriptor: | COLD-SHOCK PROTEIN CSPB, SODIUM ION | | Authors: | Delbrueck, H, Mueller, U, Perl, D, Schmid, F.X, Heinemann, U. | | Deposit date: | 2001-02-27 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of mutant forms of the Bacillus caldolyticus cold shock protein differing in thermal stability.
J.Mol.Biol., 313, 2001
|
|
