1QZC
 
 | | Coordinates of S12, SH44, LH69 and SRL separately fitted into the cryo-EM map of EF-Tu ternary complex (GDP.Kirromycin) bound 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S12 | | Authors: | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-09-16 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Incorporation of Aminoacyl-tRNA into the Ribosome as seen by Cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
1QZD
 
 | | EF-Tu.kirromycin coordinates fitted into the cryo-EM map of EF-Tu ternary complex (GDP.Kirromycin) bound 70S ribosome | | Descriptor: | Elongation factor Tu | | Authors: | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-09-16 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Incorporation of Aminoacyl-tRNA into the Ribosome as seen by Cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
1QZB
 
 | | Coordinates of the A-site tRNA model fitted into the cryo-EM map of 70S ribosome in the pre-translocational state | | Descriptor: | Phe-tRNA | | Authors: | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-09-16 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Incorporation of Aminoacyl-tRNA into the Ribosome as seen by Cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
2AKH
 
 | | Normal mode-based flexible fitted coordinates of a non-translocating SecYEG protein-conducting channel into the cryo-EM map of a SecYEG-nascent chain-70S ribosome complex from E. coli | | Descriptor: | Preprotein translocase secE subunit, Preprotein translocase secY subunit, Protein-export membrane protein secG | | Authors: | Mitra, K.M, Schaffitzel, C, Shaikh, T, Tama, F, Jenni, S, Brooks III, C.L, Ban, N, Frank, J. | | Deposit date: | 2005-08-03 | | Release date: | 2005-11-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (14.9 Å) | | Cite: | Structure of the E. coli protein-conducting channel bound to a translating ribosome.
Nature, 438, 2005
|
|
1QZA
 
 | | Coordinates of the A/T site tRNA model fitted into the cryo-EM map of EF-Tu ternary complex (GDP.Kirromycin) bound 70S ribosome | | Descriptor: | Phe-tRNA | | Authors: | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-09-16 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Incorporation of Aminoacyl-tRNA into the Ribosome as seen by Cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
1R2X
 
 | | Coordinates of L11 with 58nts of 23S rRNA fitted into the cryo-EM map of EF-Tu ternary complex (GDP.Kirromycin) bound 70S ribosome | | Descriptor: | 50S ribosomal protein L11, 58nts of 23S rRNA | | Authors: | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-09-30 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
1PN7
 
 | | Coordinates of S12, L11 proteins and P-tRNA, from the 70S X-ray structure aligned to the 70S Cryo-EM map of E.coli ribosome | | Descriptor: | 30S ribosomal protein S12, 50S ribosomal protein L11, P-tRNA | | Authors: | Valle, M, Zavialov, A, Sengupta, J, Rawat, U, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-06-12 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (10.8 Å) | | Cite: | Locking and Unlocking of Ribosomal Motions
Cell(Cambridge,Mass.), 114, 2003
|
|
1LVU
 
 | | Crystal structure of calf spleen purine nucleoside phosphorylase in a new space group with full trimer in the asymmetric unit | | Descriptor: | 2,6-DIAMINO-(S)-9-[2-(PHOSPHONOMETHOXY)PROPYL]PURINE, CALCIUM ION, Purine nucleoside phosphorylase | | Authors: | Bzowska, A, Koellner, G, Wielgus-Kutrowska, B, Stroh, A, Raszewski, G, Holy, A, Steiner, T, Frank, J. | | Deposit date: | 2002-05-29 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of calf spleen purine nucleoside phosphorylase with two full trimers in the asymmetric unit: important implications for the mechanism of catalysis
J.Mol.Biol., 342, 2004
|
|
1PN6
 
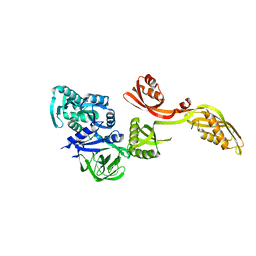 | | Domain-wise fitting of the crystal structure of T.thermophilus EF-G into the low resolution map of the release complex.Puromycin.EFG.GDPNP of E.coli 70S ribosome. | | Descriptor: | Elongation factor G | | Authors: | Valle, M, Zavialov, A, Sengupta, J, Rawat, U, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-06-12 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (10.8 Å) | | Cite: | Locking and Unlocking of Ribosomal Motions
Cell(Cambridge,Mass.), 114, 2003
|
|
1T1M
 
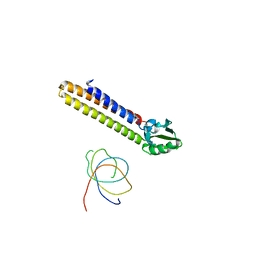 | | Binding position of ribosome recycling factor (RRF) on the E. coli 70S ribosome | | Descriptor: | 42-mer fragment of double helix from 16S rRNA, dodecamer fragment of double helix from 23S rRNA, ribosome recycling factor | | Authors: | Agrawal, R.K, Sharma, M.R, Kiel, M.C, Hirokawa, G, Booth, T.M, Spahn, C.M, Grassucci, R.A, Kaji, A, Frank, J. | | Deposit date: | 2004-04-16 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: Functional implications
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2AKI
 
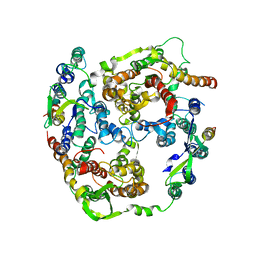 | | Normal mode-based flexible fitted coordinates of a translocating SecYEG protein-conducting channel into the cryo-EM map of a SecYEG-nascent chain-70S ribosome complex from E. coli | | Descriptor: | Preprotein translocase secE subunit, Preprotein translocase secY subunit, Protein-export membrane protein secG | | Authors: | Mitra, K, Schaffitzel, C, Shaikh, T, Tama, F, Jenni, S, Brooks III, C.L, Ban, N, Frank, J. | | Deposit date: | 2005-08-03 | | Release date: | 2005-11-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (14.9 Å) | | Cite: | Structure of the E. coli protein-conducting channel bound to a translating ribosome.
Nature, 438, 2005
|
|
1R2W
 
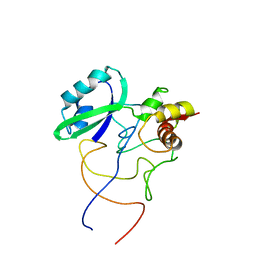 | | Coordinates of L11 with 58nts of 23S rRNA fitted into the cryo-EM map of the 70S ribosome | | Descriptor: | 50S ribosomal protein L11, 58nts of 23S rRNA | | Authors: | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-09-30 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
1ZO1
 
 | | IF2, IF1, and tRNA fitted to cryo-EM data OF E. COLI 70S initiation complex | | Descriptor: | P/I-site tRNA, translation Initiation Factor 1, translation initiation factor 2 | | Authors: | Allen, G.S, Zavialov, A, Gursky, R, Ehrenberg, M, Frank, J. | | Deposit date: | 2005-05-12 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (13.8 Å) | | Cite: | The Cryo-EM Structure of a Translation Initiation Complex from Escherichia coli.
Cell(Cambridge,Mass.), 121, 2005
|
|
1PN8
 
 | | Coordinates of S12, L11 proteins and E-site tRNA from 70S crystal structure separately fitted into the Cryo-EM map of E.coli 70S.EF-G.GDPNP complex. The atomic coordinates originally from the E-site tRNA were fitted in the position of the hybrid P/E-site tRNA. | | Descriptor: | 30S ribosomal protein S12, 50S ribosomal protein L11, E-tRNA | | Authors: | Valle, M, Zavialov, A, Sengupta, J, Rawat, U, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-06-12 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (10.8 Å) | | Cite: | Locking and Unlocking of Ribosomal Motions
Cell(Cambridge,Mass.), 114, 2003
|
|
1LV8
 
 | | Crystal structure of calf spleen purine nucleoside phosphorylase in a new space group with full trimer in the asymmetric unit | | Descriptor: | 2,6-DIAMINO-(S)-9-[2-(PHOSPHONOMETHOXY)PROPYL]PURINE, CALCIUM ION, PURINE NUCLEOSIDE PHOSPHORYLASE | | Authors: | Bzowska, A, Koellner, G, Wielgus-Kutrowska, B, Stroh, A, Raszewski, G, Holy, A, Steiner, T, Frank, J. | | Deposit date: | 2002-05-27 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of calf spleen purine nucleoside phosphorylase with two full trimers in the asymmetric unit: important implications for the mechanism of catalysis
J.Mol.Biol., 342, 2004
|
|
5T9R
 
 | | Structure of rabbit RyR1 (Ca2+-only dataset, class 3) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-09 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TB0
 
 | | Structure of rabbit RyR1 (EGTA-only dataset, all particles) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-10 | | Release date: | 2016-10-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TAS
 
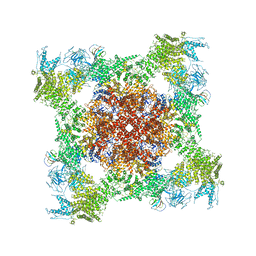 | | Structure of rabbit RyR1 (Caffeine/ATP/EGTA dataset, class 1) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-10 | | Release date: | 2016-10-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TA3
 
 | | Structure of rabbit RyR1 (Caffeine/ATP/Ca2+ dataset, class 2) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-09 | | Release date: | 2016-10-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TB2
 
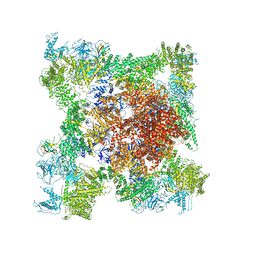 | | Structure of rabbit RyR1 (EGTA-only dataset, class 2) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-11 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TAV
 
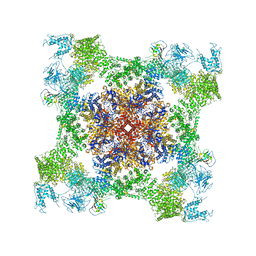 | | Structure of rabbit RyR1 (Caffeine/ATP/EGTA dataset, class 4) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-10 | | Release date: | 2016-10-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5T9N
 
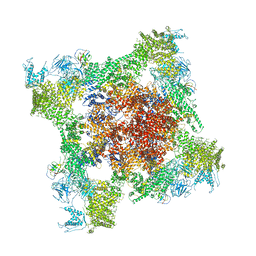 | | Structure of rabbit RyR1 (Ca2+-only dataset, class 2) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-09 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TAZ
 
 | | Structure of rabbit RyR1 (ryanodine dataset, class 3) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-10 | | Release date: | 2016-10-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TAL
 
 | | Structure of rabbit RyR1 (Caffeine/ATP/Ca2+ dataset, class 1&2) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-10 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TAW
 
 | | Structure of rabbit RyR1 (ryanodine dataset, all particles) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-10 | | Release date: | 2016-10-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
