8P89
 
 | |
8P88
 
 | |
6C8V
 
 | | X-ray structure of PqqE from Methylobacterium extorquens | | Descriptor: | Coenzyme PQQ synthesis protein E, FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER | | Authors: | Gizzi, A.S, Grove, T.L, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2018-01-25 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray and EPR Characterization of the Auxiliary Fe-S Clusters in the Radical SAM Enzyme PqqE.
Biochemistry, 57, 2018
|
|
1NAY
 
 | | GPP-Foldon:X-ray structure | | Descriptor: | Collagen-like peptide | | Authors: | Stetefeld, J. | | Deposit date: | 2002-11-29 | | Release date: | 2003-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Collagen Stabilization at Atomic Level. Crystal Structure of Designed (GlyProPro)(10)foldon
Structure, 11, 2003
|
|
5F5U
 
 | | Crystal structure of the Snu23-Prp38-MFAP1(217-258) complex of Chaetomium thermophilum | | Descriptor: | Prp38, Putative uncharacterized protein, Zinc finger domain-containing protein | | Authors: | Ulrich, A.K.C, Seeger, M, Bartlick, N, Wahl, M.C. | | Deposit date: | 2015-12-04 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.748 Å) | | Cite: | Scaffolding in the Spliceosome via Single alpha Helices.
Structure, 24, 2016
|
|
5F5V
 
 | | Crystal structure of the Snu23-Prp38-MFAP1(217-296) complex of Chaetomium thermophilum | | Descriptor: | Prp38, Putative uncharacterized protein, Zinc finger domain-containing protein | | Authors: | Ulrich, A.K.C, Seeger, M, Bartlick, N, Wahl, M.C. | | Deposit date: | 2015-12-04 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Scaffolding in the Spliceosome via Single alpha Helices.
Structure, 24, 2016
|
|
5F5S
 
 | |
5F5T
 
 | |
5GYH
 
 | |
6G70
 
 | | Structure of murine Prpf39 | | Descriptor: | Pre-mRNA-processing factor 39 | | Authors: | De Bortoli, F.D, Loll, B, Wahl, M, Heyd, F. | | Deposit date: | 2018-04-04 | | Release date: | 2019-04-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Increased versatility despite reduced molecular complexity: evolution, structure and function of metazoan splicing factor PRPF39.
Nucleic Acids Res., 47, 2019
|
|
5GY8
 
 | |
5GYC
 
 | |
5GYG
 
 | |
5GYA
 
 | |
5GV1
 
 | | Crystal structure of ENZbleach xylanase wild type | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | Chitnumsub, P, Jaruwat, A, Boonyapakorn, K, Noytanom, K. | | Deposit date: | 2016-09-01 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based protein engineering for thermostable and alkaliphilic enhancement of endo-beta-1,4-xylanase for applications in pulp bleaching
J. Biotechnol., 259, 2017
|
|
5GYB
 
 | |
5GYE
 
 | |
5GYI
 
 | |
5GY9
 
 | |
5GYF
 
 | |
4ZH3
 
 | |
4ZH4
 
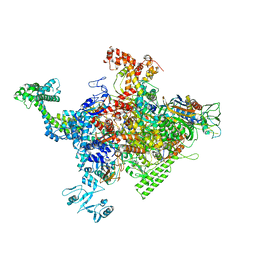 | | Crystal structure of Escherichia coli RNA polymerase in complex with CBRP18 | | Descriptor: | 5-(4-fluorophenyl)-4-[4-fluoro-3-(trifluoromethyl)phenyl]-1H-pyrazole, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Feng, Y, Ebright, R.H. | | Deposit date: | 2015-04-24 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.993 Å) | | Cite: | Structural Basis of Transcription Inhibition by CBR Hydroxamidines and CBR Pyrazoles.
Structure, 23, 2015
|
|
4BBJ
 
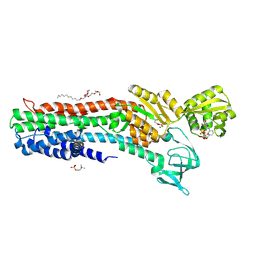 | | Copper-transporting PIB-ATPase in complex with beryllium fluoride representing the E2P state | | Descriptor: | COPPER EFFLUX ATPASE, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Mattle, D, Gourdon, P, Nissen, P. | | Deposit date: | 2012-09-25 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Copper-Transporting P-Type Atpases Use a Unique Ion-Release Pathway
Nat.Struct.Mol.Biol., 21, 2014
|
|
4ZH2
 
 | |
1AVY
 
 | | FIBRITIN DELETION MUTANT M (BACTERIOPHAGE T4) | | Descriptor: | FIBRITIN | | Authors: | Strelkov, S.V, Tao, Y, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 1997-09-22 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of bacteriophage T4 fibritin M: a troublesome packing arrangement.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
