1DSS
 
 | |
8HO1
 
 | | Crystal structure of cytochrome P450 NasF5053 mutant F387G | | Descriptor: | CALCIUM ION, Cytochrome P450-F5053, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ma, B.D, Tian, W, Qu, X, Kong, X.D. | | Deposit date: | 2022-12-09 | | Release date: | 2023-04-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering the Substrate Specificity of a P450 Dimerase Enables the Collective Biosynthesis of Heterodimeric Tryptophan-Containing Diketopiperazines.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HNZ
 
 | | Crystal structure of cytochrome P450 NasF5053 mutant E73S complexed with 6FCWP | | Descriptor: | (3~{S},8~{a}~{S})-3-[(5-fluoranyl-1~{H}-indol-3-yl)methyl]-2,3,6,7,8,8~{a}-hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, CALCIUM ION, Cytochrome P450-F5053, ... | | Authors: | Ma, B.D, Tian, W, Qu, X, Kong, X.D. | | Deposit date: | 2022-12-09 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering the Substrate Specificity of a P450 Dimerase Enables the Collective Biosynthesis of Heterodimeric Tryptophan-Containing Diketopiperazines.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HO0
 
 | | Crystal structure of cytochrome P450 NasF5053 mutant E73S complexed with 8FCWP | | Descriptor: | (3~{S},8~{a}~{S})-3-[(7-fluoranyl-1~{H}-indol-3-yl)methyl]-2,3,6,7,8,8~{a}-hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, CALCIUM ION, Cytochrome P450-F5053, ... | | Authors: | Ma, B.D, Tian, W, Qu, X, Kong, X.D. | | Deposit date: | 2022-12-09 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Engineering the Substrate Specificity of a P450 Dimerase Enables the Collective Biosynthesis of Heterodimeric Tryptophan-Containing Diketopiperazines.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HNY
 
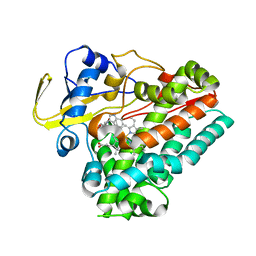 | | Crystal structure of cytochrome P450 NasF5053 mutant E73S complexed with 5FCWP | | Descriptor: | (3~{S},8~{a}~{S})-3-[(4-fluoranyl-1~{H}-indol-3-yl)methyl]-2,3,6,7,8,8~{a}-hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, Cytochrome P450-F5053, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ma, B.D, Tian, W, Qu, X, Kong, X.D. | | Deposit date: | 2022-12-09 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineering the Substrate Specificity of a P450 Dimerase Enables the Collective Biosynthesis of Heterodimeric Tryptophan-Containing Diketopiperazines.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
4ZRI
 
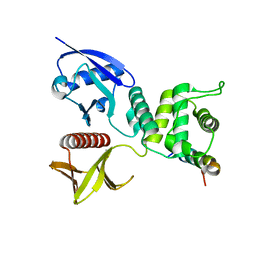 | | Crystal structure of Merlin-FERM and Lats2 | | Descriptor: | Merlin, Serine/threonine-protein kinase LATS2 | | Authors: | Li, F, Zhou, H, Long, J, Shen, Y. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway
Cell Res., 25, 2015
|
|
5JJU
 
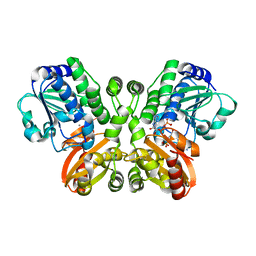 | | Crystal structure of Rv2837c complexed with 5'-pApA and 5'-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MANGANESE (II) ION, RNA (5'-R(P*AP*A)-3'), ... | | Authors: | Wang, F, He, Q, Liu, S, Gu, L. | | Deposit date: | 2016-04-25 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.312 Å) | | Cite: | Structural and biochemical insight into the mechanism of Rv2837c from Mycobacterium tuberculosis as a c-di-NMP phosphodiesterase
J.Biol.Chem., 291, 2016
|
|
7V2Z
 
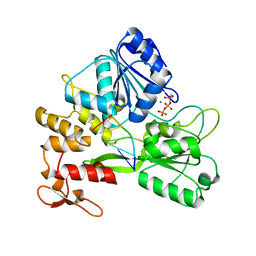 | | ZIKV NS3helicase in complex with ssRNA and ATP-Mn2+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Core protein, MANGANESE (II) ION, ... | | Authors: | Lin, M.M, Yang, H.T. | | Deposit date: | 2021-08-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.10101676 Å) | | Cite: | Structural Basis of Zika Virus Helicase in RNA Unwinding and ATP Hydrolysis.
Acs Infect Dis., 8, 2022
|
|
6O6W
 
 | | Solution structure of human myeloid-derived growth factor | | Descriptor: | Myeloid-derived growth factor | | Authors: | Bortnov, V, Tonelli, M, Lee, W, Markley, J.L, Mosher, D.F. | | Deposit date: | 2019-03-07 | | Release date: | 2019-11-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human myeloid-derived growth factor suggests a conserved function in the endoplasmic reticulum.
Nat Commun, 10, 2019
|
|
7E12
 
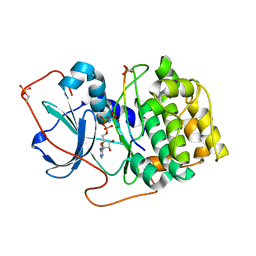 | | Crystal structure of PKAc-A11E complex | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, THR-ARG-SER-GLU-ILE-ARG-ARG-ALA-SER-THR-ILE-GLU, ... | | Authors: | Qin, J, Lin, L, Yuchi, Z. | | Deposit date: | 2021-01-28 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Structures of PKA-phospholamban complexes reveal a mechanism of familial dilated cardiomyopathy.
Elife, 11, 2022
|
|
7E11
 
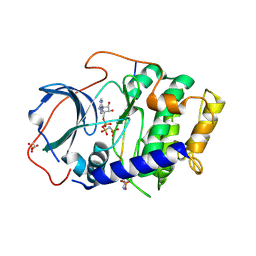 | | Crystal structure of PKAc-PLN R9C complex | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PLN, ... | | Authors: | Qin, J, Lin, L, Yuchi, Z. | | Deposit date: | 2021-01-28 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | Structures of PKA-phospholamban complexes reveal a mechanism of familial dilated cardiomyopathy.
Elife, 11, 2022
|
|
7E0Z
 
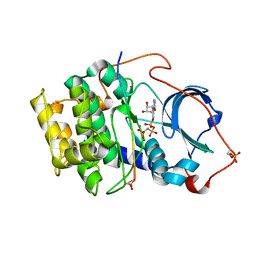 | | Crystal structure of PKAc-PLN complex | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PLN, ... | | Authors: | Qin, J, Yuchi, Z. | | Deposit date: | 2021-01-28 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Structures of PKA-phospholamban complexes reveal a mechanism of familial dilated cardiomyopathy.
Elife, 11, 2022
|
|
5K7K
 
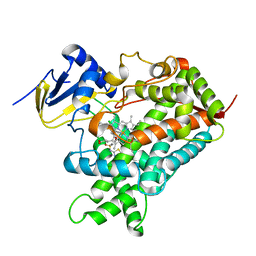 | |
6LPW
 
 | |
6LPV
 
 | |
5FO9
 
 | | Crystal Structure of Human Complement C3b in Complex with CR1 (CCP15- 17) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COMPLEMENT C3 BETA CHAIN, COMPLEMENT C3B ALPHA' CHAIN, ... | | Authors: | Forneris, F, Wu, J, Xue, X, Gros, P. | | Deposit date: | 2015-11-18 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Regulators of Complement Activity Mediate Inhibitory Mechanisms Through a Common C3B-Binding Mode.
Embo J., 35, 2016
|
|
5FO8
 
 | | Crystal Structure of Human Complement C3b in Complex with MCP (CCP1-4) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COMPLEMENT C3, ... | | Authors: | Forneris, F, Wu, J, Xue, X, Gros, P. | | Deposit date: | 2015-11-18 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulators of Complement Activity Mediate Inhibitory Mechanisms Through a Common C3B-Binding Mode.
Embo J., 35, 2016
|
|
5FO7
 
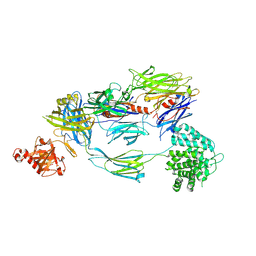 | | Crystal Structure of Human Complement C3b at 2.8 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COMPLEMENT C3 BETA CHAIN, COMPLEMENT C3B ALPHA' CHAIN | | Authors: | Forneris, F, Wu, J, Xue, X, Gros, P. | | Deposit date: | 2015-11-18 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Regulators of Complement Activity Mediate Inhibitory Mechanisms Through a Common C3B-Binding Mode.
Embo J., 35, 2016
|
|
5YJH
 
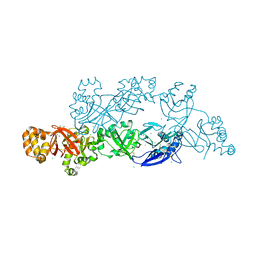 | | Structural insights into periostin functions | | Descriptor: | CALCIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Liu, H, Liu, J, Xu, F. | | Deposit date: | 2017-10-10 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.957 Å) | | Cite: | Structural characterizations of human periostin dimerization and cysteinylation.
FEBS Lett., 592, 2018
|
|
5YJG
 
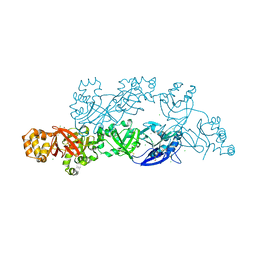 | | Structural insights into periostin functions | | Descriptor: | CALCIUM ION, CHLORIDE ION, CYSTEINE, ... | | Authors: | Liu, H, Liu, J, Xu, F. | | Deposit date: | 2017-10-10 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural characterizations of human periostin dimerization and cysteinylation.
FEBS Lett., 592, 2018
|
|
5FOA
 
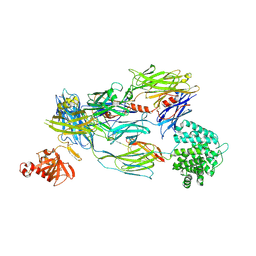 | | Crystal Structure of Human Complement C3b in complex with DAF (CCP2-4) | | Descriptor: | COMPLEMENT C3 BETA CHAIN, COMPLEMENT C3B ALPHA CHAIN, DECAY ACCELERATING FACTOR, ... | | Authors: | Forneris, F, Wu, J, Xue, X, Gros, P. | | Deposit date: | 2015-11-18 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.188 Å) | | Cite: | Regulators of Complement Activity Mediate Inhibitory Mechanisms Through a Common C3B-Binding Mode.
Embo J., 35, 2016
|
|
5FOB
 
 | | Crystal Structure of Human Complement C3b in complex with Smallpox Inhibitor of Complement (SPICE) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COMPLEMENT C3 BETA CHAIN, ... | | Authors: | Forneris, F, Wu, J, Xue, X, Gros, P. | | Deposit date: | 2015-11-18 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Regulators of Complement Activity Mediate Inhibitory Mechanisms Through a Common C3B-Binding Mode.
Embo J., 35, 2016
|
|
8J5U
 
 | | Crystal structure of Mycobacterium tuberculosis OppA complexed with an endogenous oligopeptide | | Descriptor: | Endogenous oligopeptide, Uncharacterized protein Rv1280c | | Authors: | Yang, X, Hu, T, Zhang, B, Rao, Z. | | Deposit date: | 2023-04-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | An oligopeptide permease, OppABCD, requires an iron-sulfur cluster domain for functionality.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8J5S
 
 | | Cryo-EM structure of Mycobacterium tuberculosis OppABCD in the pre-catalytic intermediate state | | Descriptor: | Endogenous oligopeptide, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Yang, X, Hu, T, Zhang, B, Rao, Z. | | Deposit date: | 2023-04-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | An oligopeptide permease, OppABCD, requires an iron-sulfur cluster domain for functionality.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8J5R
 
 | | Cryo-EM structure of Mycobacterium tuberculosis OppABCD in the resting state | | Descriptor: | IRON/SULFUR CLUSTER, Putative peptide transport permease protein Rv1282c, Putative peptide transport permease protein Rv1283c, ... | | Authors: | Yang, X, Hu, T, Zhang, B, Rao, Z. | | Deposit date: | 2023-04-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | An oligopeptide permease, OppABCD, requires an iron-sulfur cluster domain for functionality.
Nat.Struct.Mol.Biol., 31, 2024
|
|
