1ZXF
 
 | | Solution structure of a self-sacrificing resistance protein, CalC from Micromonospora echinospora | | Descriptor: | CalC | | Authors: | Singh, S, Hager, M.H, Zhang, C, Griffith, B.R, Lee, M.S, Hallenga, K, Markley, J.L, Thorson, J.S, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-06-08 | | Release date: | 2005-12-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the self-sacrifice mechanism of enediyne resistance.
Acs Chem.Biol., 1, 2006
|
|
2KOM
 
 | | Solution structure of humar Par-3b PDZ2 (residues 451-549) | | Descriptor: | Partitioning defective 3 homolog | | Authors: | Volkman, B.F, Tyler, R.C, Peterson, F.C, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-09-24 | | Release date: | 2009-11-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Rapid, robotic, small-scale protein production for NMR screening and structure determination.
Protein Sci., 19, 2010
|
|
2KLI
 
 | | Structural Basis for the Photoconversion of A Phytochrome to the Activated FAR-RED LIGHT-ABSORBING Form | | Descriptor: | PHYCOCYANOBILIN, Sensor protein | | Authors: | Cornilescu, C.C, Cornilescu, G, Ulijasz, A.T, Zhang, J, Rivera, M, Vierstra, R.D, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-07-03 | | Release date: | 2009-11-03 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the photoconversion of a phytochrome to the activated Pfr form
Nature, 463, 2010
|
|
2A33
 
 | | X-Ray Structure of a Lysine Decarboxylase-Like Protein from Arabidopsis Thaliana Gene AT2G37210 | | Descriptor: | MAGNESIUM ION, SULFATE ION, hypothetical protein | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Mccoy, J.G, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-06-23 | | Release date: | 2005-07-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray crystal structures of the conserved hypothetical proteins from Arabidopsis thaliana gene loci At5g11950 and AT2g37210.
Proteins, 65, 2006
|
|
2GOW
 
 | | Solution structure of BC059385 from Homo sapiens | | Descriptor: | Ubiquitin-like protein 3 | | Authors: | Volkman, B.F, de la Cruz, N.B, Lytle, B.L, Peterson, F.C, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-04-14 | | Release date: | 2006-04-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a membrane-anchored ubiquitin-fold (MUB) protein from Homo sapiens.
Protein Sci., 16, 2007
|
|
2KOI
 
 | | Refined solution structure of a cyanobacterial phytochrome GAF domain in the red light-absorbing ground state | | Descriptor: | PHYCOCYANOBILIN, Sensor protein | | Authors: | Cornilescu, C.C, Cornilescu, G, Ulijasz, A.T, Vierstra, R.D, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-09-22 | | Release date: | 2009-11-03 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the photoconversion of a phytochrome to the activated Pfr form
Nature, 463, 2010
|
|
2ATF
 
 | | X-RAY STRUCTURE OF cysteine dioxygenase type I FROM MUS MUSCULUS MM.241056 | | Descriptor: | 1,2-ETHANEDIOL, Cysteine dioxygenase type I, NICKEL (II) ION | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Mccoy, J.G, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-08-24 | | Release date: | 2005-10-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and mechanism of mouse cysteine dioxygenase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1XO8
 
 | | Solution structure of AT1g01470 from Arabidopsis Thaliana | | Descriptor: | At1g01470 | | Authors: | Singh, S, Cornilescu, C.C, Tyler, R.C, Cornilescu, G, Tonelli, M, Lee, M.S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-10-06 | | Release date: | 2004-10-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a late embryogenesis abundant protein (LEA14) from Arabidopsis thaliana, a cellular stress-related protein
Protein Sci., 14, 2005
|
|
1XFL
 
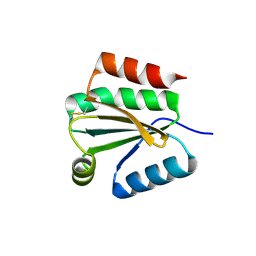 | | Solution Structure of Thioredoxin h1 from Arabidopsis Thaliana | | Descriptor: | Thioredoxin h1 | | Authors: | Peterson, F.C, Lytle, B.L, Sampath, S, Vinarov, D, Tyler, E, Shahan, M, Markley, J.L, Volkman, B.F, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-09-15 | | Release date: | 2004-09-28 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of thioredoxin h1 from Arabidopsis thaliana.
Protein Sci., 14, 2005
|
|
2I9Y
 
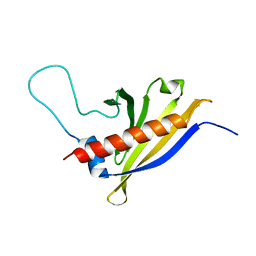 | | Solution structure of Arabidopsis thaliana protein At1g70830, a member of the major latex protein family | | Descriptor: | major latex protein-like protein 28 or MLP-like protein 28 | | Authors: | Volkman, B.F, de la Cruz, N.B, Lytle, B.L, Peterson, F.C, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-09-06 | | Release date: | 2006-09-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structures of two Arabidopsis thaliana major latex proteins represent novel helix-grip folds.
Proteins, 76, 2009
|
|
1XFI
 
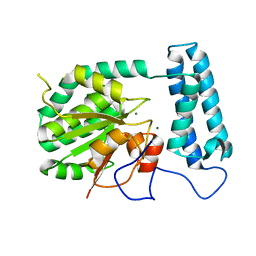 | | X-ray structure of gene product from Arabidopsis thaliana At2g17340 | | Descriptor: | MAGNESIUM ION, unknown protein | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-09-14 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure at 1.7 A resolution of the protein product of the At2g17340 gene from Arabidopsis thaliana.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
2L65
 
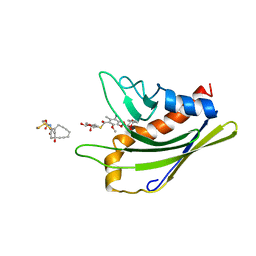 | | HADDOCK calculated model of the complex of the resistance protein CalC and Calicheamicin-Gamma | | Descriptor: | 2,4-dideoxy-4-(ethylamino)-3-O-methyl-alpha-L-threo-pentopyranose-(1-2)-4-amino-4,6-dideoxy-beta-D-glucopyranose, 2,6-dideoxy-4-thio-beta-D-allopyranose, 3-O-methyl-alpha-L-rhamnopyranose, ... | | Authors: | Singh, S, Markley, J.L, Thorson, J.S, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-11-15 | | Release date: | 2011-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the self-sacrifice mechanism of enediyne resistance.
Acs Chem.Biol., 1, 2006
|
|
1XOY
 
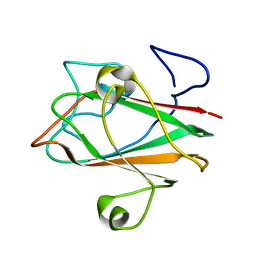 | | Solution structure of At3g04780.1, an Arabidopsis ortholog of the C-terminal domain of human thioredoxin-like protein | | Descriptor: | hypothetical protein At3g04780.1 | | Authors: | Song, J, Robert, C.T, Lee, M.S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-10-07 | | Release date: | 2004-10-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of At3g04780.1-des15, an Arabidopsis thaliana ortholog of the C-terminal domain of human thioredoxin-like protein.
Protein Sci., 14, 2005
|
|
2JZ4
 
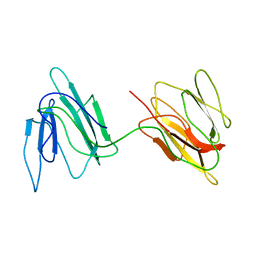 | | Putative 32 kDa myrosinase binding protein At3g16450.1 from Arabidopsis thaliana | | Descriptor: | Jasmonate inducible protein isolog | | Authors: | Takeda, N, Sugimori, N, Torizawa, T, Terauchi, T, Ono, A.M, Yagi, H, Yamaguchi, Y, Kato, K, Ikeya, T, Guntert, P, Aceti, D.J, Markley, J.L, Kainosho, M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-12-28 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of the putative 32 kDa myrosinase-binding protein from Arabidopsis (At3g16450.1) determined by SAIL-NMR.
Febs J., 275, 2008
|
|
2IFU
 
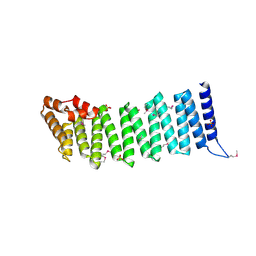 | | Crystal Structure of a Gamma-SNAP from Danio rerio | | Descriptor: | SULFATE ION, gamma-snap | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Mccoy, J.G, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-09-21 | | Release date: | 2006-10-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and dynamics of gamma-SNAP: insight into flexibility of proteins from the SNAP family.
Proteins, 70, 2008
|
|
2IL4
 
 | | Crystal structure of At1g77540-Coenzyme A Complex | | Descriptor: | COENZYME A, Protein At1g77540 | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-10-02 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Structure of Arabidopsis thaliana At1g77540 Protein, a Minimal Acetyltransferase from the COG2388 Family.
Biochemistry, 45, 2006
|
|
1XMB
 
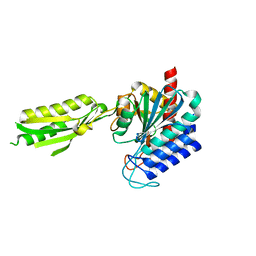 | | X-ray structure of IAA-aminoacid hydrolase from Arabidopsis thaliana gene AT5G56660 | | Descriptor: | IAA-amino acid hydrolase homolog 2 | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-10-01 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of ILL2, an auxin-conjugate amidohydrolase from Arabidopsis thaliana.
Proteins, 74, 2009
|
|
2I5T
 
 | | Crystal Structure of hypothetical protein LOC79017 from Homo sapiens | | Descriptor: | Protein C7orf24 | | Authors: | Bae, E, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-08-25 | | Release date: | 2006-09-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of Homo sapiens protein LOC79017.
Proteins, 70, 2008
|
|
1Z7X
 
 | | X-ray structure of human ribonuclease inhibitor complexed with ribonuclease I | | Descriptor: | CITRIC ACID, Ribonuclease I, Ribonuclease inhibitor | | Authors: | McCoy, J.G, Johnson, R.J, Raines, R.T, Bitto, E, Bingman, C.A, Wesenberg, G.E, Allard, S.T.M, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-03-28 | | Release date: | 2005-06-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Inhibition of human pancreatic ribonuclease by the human ribonuclease inhibitor protein.
J.Mol.Biol., 368, 2007
|
|
2KOJ
 
 | |
1Z90
 
 | | X-ray structure of gene product from arabidopsis thaliana at3g03250, a putative UDP-glucose pyrophosphorylase | | Descriptor: | AT3g03250 protein | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-03-31 | | Release date: | 2005-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure and Dynamics of UDP-Glucose Pyrophosphorylase from Arabidopsis thaliana with Bound UDP-Glucose and UTP.
J.Mol.Biol., 366, 2007
|
|
2A3L
 
 | | X-Ray Structure of Adenosine 5'-Monophosphate Deaminase from Arabidopsis Thaliana in Complex with Coformycin 5'-Phosphate | | Descriptor: | AMP deaminase, COFORMYCIN 5'-PHOSPHATE, PHOSPHATE ION, ... | | Authors: | Han, B.W, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-06-25 | | Release date: | 2005-07-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Membrane association, mechanism of action, and structure of Arabidopsis embryonic factor 1 (FAC1).
J.Biol.Chem., 281, 2006
|
|
2KMW
 
 | |
2GMK
 
 | | Crystal structure of onconase double mutant with spontaneously-assembled (AMP) 4 stack | | Descriptor: | ADENOSINE MONOPHOSPHATE, P-30 protein | | Authors: | Bae, E, Lee, J.E, Raines, R.T, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-04-06 | | Release date: | 2006-04-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for catalysis by onconase.
J.Mol.Biol., 375, 2008
|
|
2GW6
 
 | |
