3S82
 
 | |
3K2E
 
 | |
5JTK
 
 | |
3SDS
 
 | |
9NPY
 
 | |
9NPX
 
 | |
8CSO
 
 | |
8CU5
 
 | |
8CU9
 
 | |
8CTR
 
 | |
8D1X
 
 | |
8D57
 
 | |
8D2Z
 
 | |
9AV7
 
 | |
9AV6
 
 | |
9B0M
 
 | |
7RCR
 
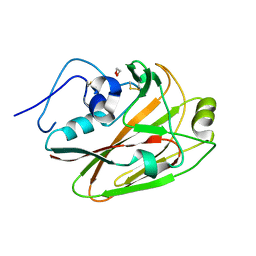 | |
7RBX
 
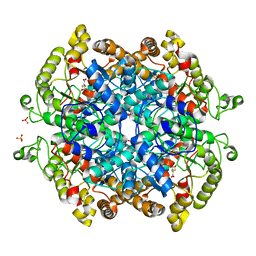 | | Crystal structure of isocitrate lyase and phosphorylmutase:isocitrate lyase from Brucella melitensis biovar Abortus 2308 bound to itaconic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-methylidenebutanedioic acid, Isocitrase, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Edwards, T.E, Abendroth, J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2021-07-06 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Aconitate decarboxylase 1 participates in the control of pulmonary Brucella infection in mice.
Plos Pathog., 17, 2021
|
|
7V0H
 
 | |
8DGD
 
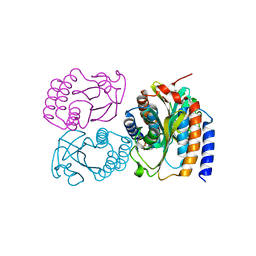 | | Crystal Structure of GAF domain-containing protein, from Klebsiella pneumoniae | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GAF domain-containing protein, ZINC ION | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Delker, S.L, Weiss, M.J, Lorimer, D.D, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-06-23 | | Release date: | 2023-03-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of GAF domain-containing protein, from Klebsiella pneumoniae
to be published
|
|
6WNG
 
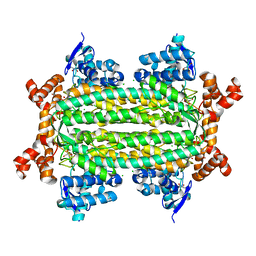 | |
7UG3
 
 | |
7U0O
 
 | |
9ASD
 
 | | VIR-7229 Fab fragment bound the SARS-CoV-2 BA.2.86 spike trimer (local refinement of the BA 2.86 RBD/VIR-7229 VHVL) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, VIR-7229 Fab heavy chain, ... | | Authors: | Park, Y.J, Tortorici, M.A, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2024-02-25 | | Release date: | 2024-10-16 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A potent pan-sarbecovirus neutralizing antibody resilient to epitope diversification.
Cell, 187, 2024
|
|
9ATM
 
 | | SARS-CoV-2 EG.5 RBD bound to the VIR-7229 and the S2H97 Fab fragments | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rietz, T, Park, Y.J, Errico, J, Czudnochowski, N, Nix, J.C, Corti, D, Snell, G, Marco, A.D, Pinto, D, Cameroni, E, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2024-02-27 | | Release date: | 2024-10-16 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A potent pan-sarbecovirus neutralizing antibody resilient to epitope diversification.
Cell, 187, 2024
|
|
