7BQ1
 
 | | X-ray structure of human PPARalpha ligand binding domain-intrinsic fatty acid (E. coli origin)-SRC1 coactivator peptide co-crystals obtained by co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, GLYCEROL, PALMITIC ACID, ... | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | Deposit date: | 2020-03-23 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
4JF6
 
 | | Structure of OXA-23 at pH 7.0 | | Descriptor: | Beta-lactamase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2013-02-27 | | Release date: | 2013-09-25 | | Last modified: | 2013-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Carbapenemase Activity of the OXA-23 beta-Lactamase from Acinetobacter baumannii.
Chem.Biol., 20, 2013
|
|
7BPY
 
 | | X-ray structure of human PPARalpha ligand binding domain-clofibric acid-SRC1 coactivator peptide co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 2-(4-chloranylphenoxy)-2-methyl-propanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | Deposit date: | 2020-03-23 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
7BQ2
 
 | | X-ray structure of human PPARalpha ligand binding domain-pemafibrate-SRC1 coactivator peptide co-crystals obtained by soaking | | Descriptor: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, 15-meric peptide from Nuclear receptor coactivator 1, GLYCEROL, ... | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | Deposit date: | 2020-03-23 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
7EGK
 
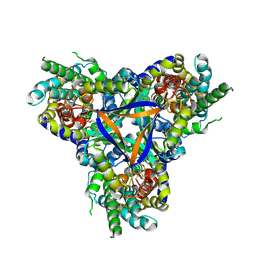 | | Bicarbonate transporter complex SbtA-SbtB bound to AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Membrane-associated protein SbtB, SODIUM ION, ... | | Authors: | Fang, S, Huang, X, Zhang, X, Zhang, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular mechanism underlying transport and allosteric inhibition of bicarbonate transporter SbtA.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EGL
 
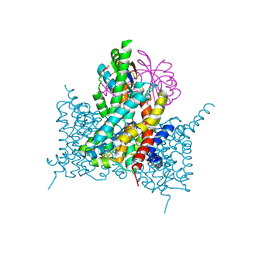 | | Bicarbonate transporter complex SbtA-SbtB bound to HCO3- | | Descriptor: | BICARBONATE ION, Membrane-associated protein SbtB, SODIUM ION, ... | | Authors: | Fang, S, Huang, X, Zhang, X, Zhang, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular mechanism underlying transport and allosteric inhibition of bicarbonate transporter SbtA.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8VEW
 
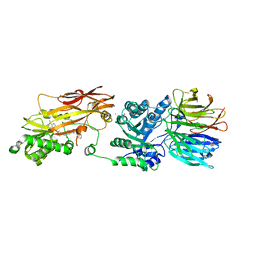 | | Crystal structure of PRMT5:MEP50 in complex with MTA and oxamide compound 24 | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, 5-{2-[(2R,5S)-5-methyl-2-phenylpiperidin-1-yl](oxo)acetamido}pyridine-3-carboxamide, CHLORIDE ION, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2023-12-20 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Discovery of TNG908: A Selective, Brain Penetrant, MTA-Cooperative PRMT5 Inhibitor That Is Synthetically Lethal with MTAP -Deleted Cancers.
J.Med.Chem., 67, 2024
|
|
8VEO
 
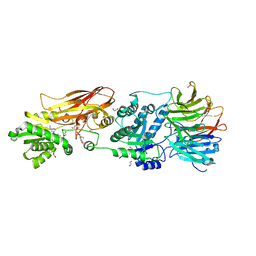 | | Crystal structure of PRMT5:MEP50 in complex with MTA | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2023-12-20 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery of TNG908: A Selective, Brain Penetrant, MTA-Cooperative PRMT5 Inhibitor That Is Synthetically Lethal with MTAP -Deleted Cancers.
J.Med.Chem., 67, 2024
|
|
8P4X
 
 | | FAD_ox bound dark state structure of PdLCry | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, Putative light-receptive cryptochrome (Fragment) | | Authors: | Behrmann, E, Behrmann, H. | | Deposit date: | 2023-05-23 | | Release date: | 2023-11-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | A marine cryptochrome with an inverse photo-oligomerization mechanism.
Nat Commun, 14, 2023
|
|
7EEI
 
 | |
7EA7
 
 | | crystal structure of NAP1 LIR in complex with GABARAP | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein, NAP1_LIR motif | | Authors: | Fu, T, Pan, L. | | Deposit date: | 2021-03-06 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural and biochemical advances on the recruitment of the autophagy-initiating ULK and TBK1 complexes by autophagy receptor NDP52.
Sci Adv, 7, 2021
|
|
7EA2
 
 | | crystal structure of NAP1 FIR in complex with RB1CC1 Claw domain | | Descriptor: | 5-azacytidine-induced protein 2,RB1-inducible coiled-coil protein 1, CITRATE ANION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fu, T, Pan, L. | | Deposit date: | 2021-03-06 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural and biochemical advances on the recruitment of the autophagy-initiating ULK and TBK1 complexes by autophagy receptor NDP52.
Sci Adv, 7, 2021
|
|
7EAA
 
 | | crystal structure of NDP52 SKICH domain in complex with RB1CC1 coiled-coil domain | | Descriptor: | Calcium-binding and coiled-coil domain-containing protein 2, RB1-inducible coiled-coil protein 1 | | Authors: | Fu, T, Pan, L. | | Deposit date: | 2021-03-06 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical advances on the recruitment of the autophagy-initiating ULK and TBK1 complexes by autophagy receptor NDP52.
Sci Adv, 7, 2021
|
|
6EKA
 
 | | Solid-state MAS NMR structure of the HELLF prion amyloid fibrils | | Descriptor: | Podospora anserina S mat+ genomic DNA chromosome 3, supercontig 2 | | Authors: | Martinez, D, Daskalov, A, Andreas, L, Bardiaux, B, Coustou, V, Stanek, J, Berbon, M, Noubhani, M, Kauffmann, B, Wall, J.S, Pintacuda, G, Saupe, S.J, Habenstein, B, Loquet, A. | | Deposit date: | 2017-09-25 | | Release date: | 2018-10-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Structural and molecular basis of cross-seeding barriers in amyloids
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7F5W
 
 | |
3EOB
 
 | |
4I6P
 
 | | Crystal structure of Par3-NTD domain | | Descriptor: | Partitioning defective 3 homolog | | Authors: | Wang, W, Gao, F, Gong, W, Sun, F, Feng, W. | | Deposit date: | 2012-11-29 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the intrinsic self-assembly of par-3 N-terminal domain.
Structure, 21, 2013
|
|
3EOA
 
 | |
3EO9
 
 | | Crystal structure the Fab fragment of Efalizumab | | Descriptor: | Efalizumab Fab fragment, heavy chain, light chain | | Authors: | Li, S, Ding, J. | | Deposit date: | 2008-09-26 | | Release date: | 2009-04-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Efalizumab binding to the LFA-1 alphaL I domain blocks ICAM-1 binding via steric hindrance.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6Z8R
 
 | | Copper transporter OprC | | Descriptor: | COPPER (II) ION, Putative copper transport outer membrane porin OprC | | Authors: | Bhamidimarri, S.P, van den Berg, B. | | Deposit date: | 2020-06-02 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Acquisition of ionic copper by the bacterial outer membrane protein OprC through a novel binding site.
Plos Biol., 19, 2021
|
|
6Z8U
 
 | | Copper transporter OprC | | Descriptor: | Putative copper transport outer membrane porin OprC, SILVER ION | | Authors: | Bhamidimarri, S.P, van den Berg, B. | | Deposit date: | 2020-06-02 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Acquisition of ionic copper by the bacterial outer membrane protein OprC through a novel binding site.
Plos Biol., 19, 2021
|
|
6Z8S
 
 | | Copper transporter OprC | | Descriptor: | COPPER (I) ION, Putative copper transport outer membrane porin OprC | | Authors: | Bhamidimarri, S.P, van den Berg, B. | | Deposit date: | 2020-06-02 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Acquisition of ionic copper by the bacterial outer membrane protein OprC through a novel binding site.
Plos Biol., 19, 2021
|
|
6Z8Q
 
 | | Copper transporter OprC | | Descriptor: | Putative copper transport outer membrane porin OprC, SILVER ION | | Authors: | Bhamidimarri, S.P, van den Berg, B. | | Deposit date: | 2020-06-02 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Acquisition of ionic copper by the bacterial outer membrane protein OprC through a novel binding site.
Plos Biol., 19, 2021
|
|
6Z8Z
 
 | | Copper transporter OprC | | Descriptor: | Putative copper transport outer membrane porin OprC | | Authors: | Bhamidimarri, S.P, van den Berg, B. | | Deposit date: | 2020-06-02 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Acquisition of ionic copper by the bacterial outer membrane protein OprC through a novel binding site.
Plos Biol., 19, 2021
|
|
6Z8T
 
 | | Copper transporter OprC | | Descriptor: | Putative copper transport outer membrane porin OprC, SILVER ION | | Authors: | Bhamidimarri, S.P, van den Berg, B. | | Deposit date: | 2020-06-02 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Acquisition of ionic copper by the bacterial outer membrane protein OprC through a novel binding site.
Plos Biol., 19, 2021
|
|
