7TXQ
 
 | | x-ray structure of the VioB N-acetyltransferase from Acinetobacter baumannii in the present of TDP and Acetyl-CoenzymeA | | Descriptor: | ACETYL COENZYME *A, SODIUM ION, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Herkert, N.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and function of an N-acetyltransferase from the human pathogen Acinetobacter baumannii isolate BAL_212.
Proteins, 90, 2022
|
|
7TXP
 
 | | X-ray structure of the VioB N-acetyltransferase from Acinetobacter baumannii in complex with TDP-4-amino-4,6-dideoxy-D-glucose | | Descriptor: | SODIUM ION, VioB, dTDP-4-amino-4,6-dideoxyglucose | | Authors: | Herkert, N.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and function of an N-acetyltransferase from the human pathogen Acinetobacter baumannii isolate BAL_212.
Proteins, 90, 2022
|
|
7US5
 
 | | X-ray crystal structure of GDP-D-glycero-D-manno-heptose 4,6-Dehydratase from Campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, GDP-D-GLYCERO-D-MANNO-HEPTOSE 4,6-DEHYDRATASE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Xiang, D.F, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-04-23 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reaction Mechanism and Three-Dimensional Structure of GDP-d-glycero-alpha-d-manno-heptose 4,6-Dehydratase from Campylobacter jejuni.
Biochemistry, 61, 2022
|
|
5BJU
 
 | | X-ray structure of the PglF dehydratase from Campylobacter jejuni in complex with UDP and NAD(H) | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Riegert, A.S, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-09-12 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Biochemical Investigation of PglF from Campylobacter jejuni Reveals a New Mechanism for a Member of the Short Chain Dehydrogenase/Reductase Superfamily.
Biochemistry, 56, 2017
|
|
5BJW
 
 | | X-ray structure of the PglF 4,6-dehydratase from campylobacter jejuni, T595S variant, in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Riegert, A.S, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-09-12 | | Release date: | 2017-11-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Investigation of PglF from Campylobacter jejuni Reveals a New Mechanism for a Member of the Short Chain Dehydrogenase/Reductase Superfamily.
Biochemistry, 56, 2017
|
|
5BJX
 
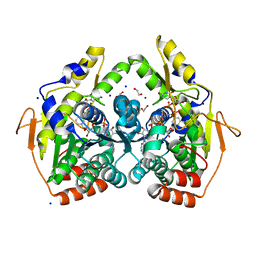 | | X-ray structure of the PglF 4,6-dehydratase from campylobacter jejuni, variant T395V, in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Riegert, A.S, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-09-12 | | Release date: | 2017-11-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Investigation of PglF from Campylobacter jejuni Reveals a New Mechanism for a Member of the Short Chain Dehydrogenase/Reductase Superfamily.
Biochemistry, 56, 2017
|
|
5BJY
 
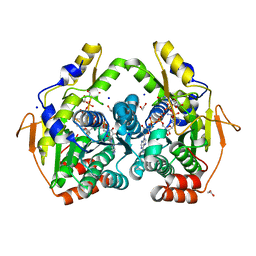 | | x-ray structure of the PglF 4,5-dehydratase from campylobacter jejuni, variant M405Y, in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Riegert, A.S, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-09-12 | | Release date: | 2017-11-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Investigation of PglF from Campylobacter jejuni Reveals a New Mechanism for a Member of the Short Chain Dehydrogenase/Reductase Superfamily.
Biochemistry, 56, 2017
|
|
5BSZ
 
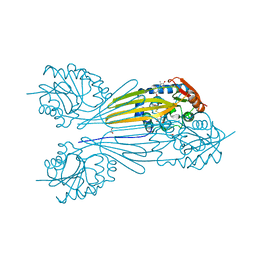 | | X-ray structure of the sugar N-methyltransferase KedS8 from Streptoalloteichus sp ATCC 53650 | | Descriptor: | 1,2-ETHANEDIOL, N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Thoden, J.B, Delvaux, N.A, Holden, H.M. | | Deposit date: | 2015-06-02 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular architecture of KedS8, a sugar N-methyltransferase from Streptoalloteichus sp. ATCC 53650.
Protein Sci., 24, 2015
|
|
2UDP
 
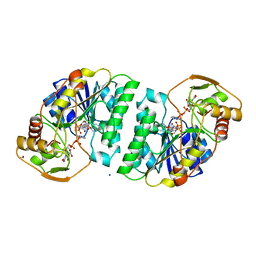 | | UDP-GALACTOSE 4-EPIMERASE COMPLEXED WITH UDP-PHENOL | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHENYL-URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Gulick, A.M, Holden, H.M. | | Deposit date: | 1997-03-08 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution X-ray structure of UDP-galactose 4-epimerase complexed with UDP-phenol.
Protein Sci., 5, 1996
|
|
8E77
 
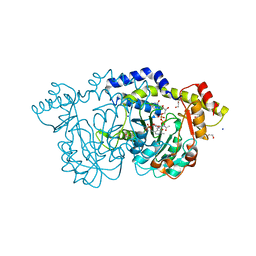 | | rystal structure of Pcryo_0616, the aminotransferase required to synthesize UDP-N-acetyl-3-amino-D-glucosaminuronic acid (UDP-GlcNAc3NA), incomplete with its external aldimine reaction intermediate | | Descriptor: | (2S,3S,4R,5R,6R)-5-(acetylamino)-6-{[(R)-{[(S)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-3-hydroxy-4-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}tetrahydro-2H-pyran-2-carboxylic acid (non-preferred name), 1,2-ETHANEDIOL, DegT/DnrJ/EryC1/StrS aminotransferase, ... | | Authors: | Hofmeister, D.L, Seltzner, C.A, Bockhaus, N.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2022-08-23 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of 2,3-diacetamido-2,3-dideoxy-d-glucuronic acid in Psychrobacter cryohalolentis K5 T.
Protein Sci., 32, 2023
|
|
8E75
 
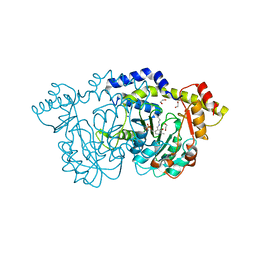 | | Crystal structure of Pcryo_0616, the aminotransferase required to synthesize UDP-N-acetyl-3-amino-D-glucosaminuronic acid (UDP-GlcNAc3NA) | | Descriptor: | 1,2-ETHANEDIOL, DegT/DnrJ/EryC1/StrS aminotransferase, SODIUM ION | | Authors: | Hofmeister, D.L, Seltzner, C.A, Bockhaus, N.J, thoden, J.B, Holden, H.M. | | Deposit date: | 2022-08-23 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of 2,3-diacetamido-2,3-dideoxy-d-glucuronic acid in Psychrobacter cryohalolentis K5 T.
Protein Sci., 32, 2023
|
|
8E62
 
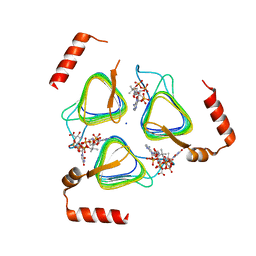 | | STRUCTURE OF Pcryo_0615 from Psychrobacter cryohalolentis, an N-acetyltransferase required to produce Diacetamido-2,3-dideoxy-D-glucuronic acid | | Descriptor: | (2S,3S,4R,5R,6R)-5-(acetylamino)-4-amino-6-{[(R)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-3-hydroxytetrahydro-2H-pyran-2-carboxylic acid, COENZYME A, SODIUM ION, ... | | Authors: | Hofmeister, D.L, Bockhaus, N.J, Seltzner, C.A, Thoden, J.B, Holden, H.M. | | Deposit date: | 2022-08-22 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of 2,3-diacetamido-2,3-dideoxy-d-glucuronic acid in Psychrobacter cryohalolentis K5 T.
Protein Sci., 32, 2023
|
|
2MYS
 
 | |
3E1K
 
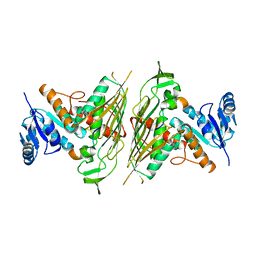 | |
8EWU
 
 | | X-ray structure of the GDP-6-deoxy-4-keto-D-lyxo-heptose-4-reductase from Campylobacter jejuni HS:15 | | Descriptor: | 1,2-ETHANEDIOL, GDP-L-fucose synthase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Xiang, D.F, Ghosh, M.K, Riegert, A.S, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-10-24 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Bifunctional Epimerase/Reductase Enzymes Facilitate the Modulation of 6-Deoxy-Heptoses Found in the Capsular Polysaccharides of Campylobacter jejuni.
Biochemistry, 62, 2023
|
|
6NBP
 
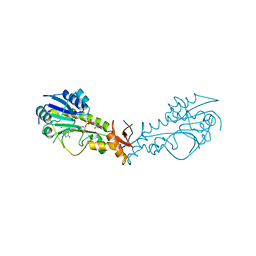 | | Crystal Structure of a Sugar N-Formyltransferase from the Plant Pathogen Pantoea ananatis | | Descriptor: | N-formyltransferase, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, PHOSPHATE ION, ... | | Authors: | Hofmeister, D.L, Thoden, J.B, Holden, H.M. | | Deposit date: | 2018-12-09 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Investigation of a sugar N-formyltransferase from the plant pathogen Pantoea ananatis.
Protein Sci., 28, 2019
|
|
6OFU
 
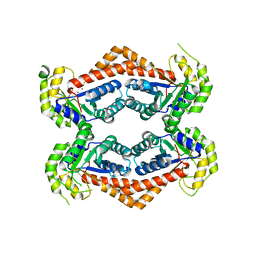 | | X-ray crystal structure of the YdjI aldolase from Escherichia coli K12 | | Descriptor: | CHLORIDE ION, YdjI aldolase, ZINC ION | | Authors: | Dopkins, B.J, Thoden, J.B, Huddleston, J.P, Narindoshvili, T, Fose, B, Rachel, F.M, Holden, H.M. | | Deposit date: | 2019-04-01 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Functional Characterization of YdjI, an Aldolase of Unknown Specificity inEscherichia coliK12.
Biochemistry, 58, 2019
|
|
6CBO
 
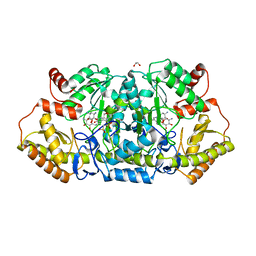 | | X-ray structure of GenB1 from micromonospora echinospora in complex with neamine and PLP (as the external aldimine) | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2-amino-2,6-dideoxy-6-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]-alpha-D-glucopyranoside, 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Dow, G.T, Thoden, J.B, Holden, H.M. | | Deposit date: | 2018-02-03 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The three-dimensional structure of NeoB: An aminotransferase involved in the biosynthesis of neomycin.
Protein Sci., 27, 2018
|
|
6CBM
 
 | | x-ray structure of NeoB from streptomyces fradiae in complex with PLP and neomycin (as the external aldimine) at pH 9 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-[(3-O-{2-amino-2,6-dideoxy-6-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]-alpha-D-glucopyranosyl}-beta-D-ribofuranosyl)oxy]-3-hydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Thoden, J.B, Dow, G.T, Holden, H.M. | | Deposit date: | 2018-02-03 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The three-dimensional structure of NeoB: An aminotransferase involved in the biosynthesis of neomycin.
Protein Sci., 27, 2018
|
|
8DB5
 
 | | Crystal structure of the GDP-D-glycero-4-keto-d-lyxo-heptose-3,5-epimerase from Campylobacter jejuni, serotype HS:15 | | Descriptor: | CHLORIDE ION, GDP-D-glycero-4-keto-d-lyxo-heptose-3,5-epimerase, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Thoden, J.B, Ghosh, M.K, Xiang, D.F, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-06-14 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | C3- and C3/C5-Epimerases Required for the Biosynthesis of the Capsular Polysaccharides from Campylobacter jejuni .
Biochemistry, 61, 2022
|
|
8DCO
 
 | | Crystal structure of the GDP-D-glycero-4-keto-D-lyxo-heptose-3,5-epimerase from Campylobacter jejuni, serotype HS:42 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GDP-D-glycero-4-keto-D-lyxo-heptose-3,5-epimerase, ... | | Authors: | Thoden, J.B, Xiang, D.F, Ghosh, M.K, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-06-17 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | C3- and C3/C5-Epimerases Required for the Biosynthesis of the Capsular Polysaccharides from Campylobacter jejuni .
Biochemistry, 61, 2022
|
|
8DCL
 
 | | Crystal structure of the GDP-D-glycero-4-keto-D-lyxo-heptose-3-epimerase from campylobacter jejuni, serotype HS:23/36 | | Descriptor: | 1,2-ETHANEDIOL, GDP-D-glycero-4-keto-D-lyxo-heptose-3-epimerase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Ghosh, M.K, Xiang, D.F, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-06-16 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | C3- and C3/C5-Epimerases Required for the Biosynthesis of the Capsular Polysaccharides from Campylobacter jejuni .
Biochemistry, 61, 2022
|
|
8DAK
 
 | | Crystal structure of the GDP-D-glycero-4-keto-d-lyxo-heptose-3-epimerase from Campylobacter jejuni, serotype HS:3 | | Descriptor: | CHLORIDE ION, GDP-D-glycero-4-keto-d-lyxo-heptose-3-epimerase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Ghosh, M.K, Xiang, D.F, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-06-13 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | C3- and C3/C5-Epimerases Required for the Biosynthesis of the Capsular Polysaccharides from Campylobacter jejuni .
Biochemistry, 61, 2022
|
|
6CBK
 
 | | X-ray structure of NeoB from Streptomyces fradiae in complex with PMP | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Neamine transaminase NeoN, ... | | Authors: | Thoden, J.B, Dow, G.T, Holden, H.M. | | Deposit date: | 2018-02-03 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The three-dimensional structure of NeoB: An aminotransferase involved in the biosynthesis of neomycin.
Protein Sci., 27, 2018
|
|
6CBL
 
 | | x-ray structure of NeoB from Streptomyces fradiae in complex with neamine as an external aldimine | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2-amino-2,6-dideoxy-6-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]-alpha-D-glucopyranoside, CHLORIDE ION, Neamine transaminase NeoN | | Authors: | Thoden, J.B, Dow, G.T, Holden, H.M. | | Deposit date: | 2018-02-03 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The three-dimensional structure of NeoB: An aminotransferase involved in the biosynthesis of neomycin.
Protein Sci., 27, 2018
|
|
