6ECU
 
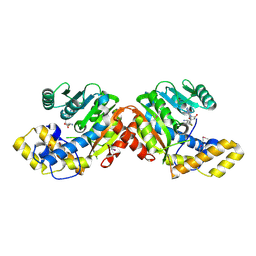 | |
4X7Y
 
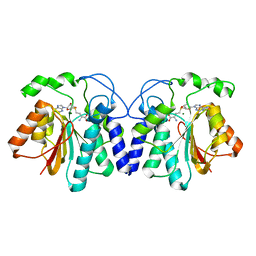 | |
6D5K
 
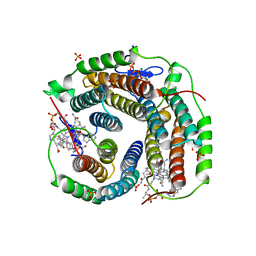 | | Structure of Human ATP:Cobalamin Adenosyltransferase bound to ATP, and Adenosylcobalamin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-DEOXYADENOSINE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Dodge, G.J, Campanello, G, Smith, J.L, Banerjee, R. | | Deposit date: | 2018-04-19 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Sacrificial Cobalt-Carbon Bond Homolysis in Coenzyme B12as a Cofactor Conservation Strategy.
J. Am. Chem. Soc., 140, 2018
|
|
7UCI
 
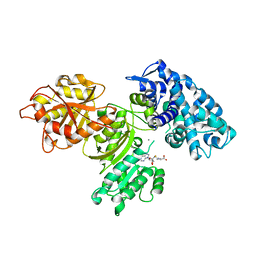 | | SxtA Methyltransferase and decarboxylase didomain in complex with Mn2+ and SAH | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Polyketide synthase-related protein, ... | | Authors: | Lao, Y, Skiba, M.A, Smith, J.L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Control of Methylation Extent in Polyketide Synthase Metal-Dependent C -Methyltransferases.
Acs Chem.Biol., 17, 2022
|
|
7UCL
 
 | |
4X7X
 
 | | MycF mycinamicin III 3'-O-methyltransferase (E35Q, E139A variant) in complex with Mg, SAH and macrocin | | Descriptor: | 2-[(4R,5S,6S,7R,9R,11E,13E,15R,16R)-6-[(2R,3R,4R,5S,6R)-4-(dimethylamino)-5-[(2S,4R,5S,6S)-4,6-dimethyl-4,5-bis(oxidanyl)oxan-2-yl]oxy-6-methyl-3-oxidanyl-oxan-2-yl]oxy-16-ethyl-15-[[(2R,3R,4R,5S,6R)-3-methoxy-6-methyl-4,5-bis(oxidanyl)oxan-2-yl]oxymethyl]-5,9,13-trimethyl-4-oxidanyl-2,10-bis(oxidanylidene)-1-oxacyclohexadeca-11,13-dien-7-yl]ethanal, MAGNESIUM ION, Mycinamicin III 3''-O-methyltransferase, ... | | Authors: | Bernard, S.M, Smith, J.L. | | Deposit date: | 2014-12-09 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Substrate Specificity and Regiochemistry in the MycF/TylF Family of Sugar O-Methyltransferases.
Acs Chem.Biol., 10, 2015
|
|
6ECX
 
 | | StiE O-MT residues 942-1257 | | Descriptor: | GLYCEROL, S-ADENOSYLMETHIONINE, StiE protein | | Authors: | Skiba, M.A, Bivins, M.B, Smith, J.L. | | Deposit date: | 2018-08-08 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Polyketide Synthase O-Methylation.
ACS Chem. Biol., 13, 2018
|
|
6ECV
 
 | | StiD O-MT residues 976-1266 | | Descriptor: | CHLORIDE ION, S-ADENOSYL-L-HOMOCYSTEINE, StiD protein | | Authors: | Skiba, M.A, Bivins, M.M, Smith, J.L. | | Deposit date: | 2018-08-08 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural Basis of Polyketide Synthase O-Methylation.
ACS Chem. Biol., 13, 2018
|
|
6ECW
 
 | | StiD O-MT residues 956-1266 | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, StiD protein | | Authors: | Skiba, M.A, Bivins, M.M, Smith, J.L. | | Deposit date: | 2018-08-08 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Polyketide Synthase O-Methylation.
ACS Chem. Biol., 13, 2018
|
|
1OX6
 
 | |
1OX4
 
 | |
1RFS
 
 | | RIESKE SOLUBLE FRAGMENT FROM SPINACH | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, RIESKE PROTEIN | | Authors: | Carrell, C.J, Zhang, H, Cramer, W.A, Smith, J.L. | | Deposit date: | 1997-08-14 | | Release date: | 1998-01-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Biological identity and diversity in photosynthesis and respiration: structure of the lumen-side domain of the chloroplast Rieske protein.
Structure, 5, 1997
|
|
1OX5
 
 | |
5THY
 
 | |
5TZ7
 
 | |
5TZ6
 
 | |
5TZ5
 
 | |
7R7G
 
 | |
7R7F
 
 | |
7R7E
 
 | |
5THZ
 
 | |
5VE5
 
 | | Crystal structure of persulfide dioxygenase rhodanese fusion protein with rhodanese domain inactivating mutation (C314S) from Burkholderia phytofirmans in complex with glutathione | | Descriptor: | BpPRF, CHLORIDE ION, FE (III) ION, ... | | Authors: | Motl, N, Skiba, M.A, Smith, J.L, Banerjee, R. | | Deposit date: | 2017-04-03 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and biochemical analyses indicate that a bacterial persulfide dioxygenase-rhodanese fusion protein functions in sulfur assimilation.
J. Biol. Chem., 292, 2017
|
|
5VE4
 
 | | Crystal structure of persulfide dioxygenase-rhodanese fusion protein with rhodanese domain inactivating mutation (C314S) from Burkholderia phytofirmans | | Descriptor: | BpPRF, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Motl, N, Skiba, M.A, Smith, J.L, Banerjee, R. | | Deposit date: | 2017-04-03 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and biochemical analyses indicate that a bacterial persulfide dioxygenase-rhodanese fusion protein functions in sulfur assimilation.
J. Biol. Chem., 292, 2017
|
|
1HCZ
 
 | | LUMEN-SIDE DOMAIN OF REDUCED CYTOCHROME F AT-35 DEGREES CELSIUS | | Descriptor: | CYTOCHROME F, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Martinez, S.E, Huang, D, Szczepaniak, A, Cramer, W.A, Smith, J.L. | | Deposit date: | 1996-09-18 | | Release date: | 1997-03-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The heme redox center of chloroplast cytochrome f is linked to a buried five-water chain.
Protein Sci., 5, 1996
|
|
5DOZ
 
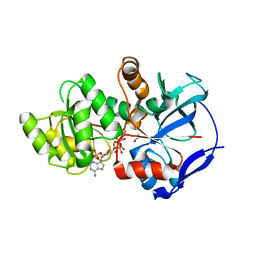 | | Crystal structure of JamJ enoyl reductase (NADPH bound) | | Descriptor: | ACETATE ION, JamJ, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Khare, D, Smith, J.L. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural Basis for Cyclopropanation by a Unique Enoyl-Acyl Carrier Protein Reductase.
Structure, 23, 2015
|
|
