8SQX
 
 | |
8SMU
 
 | | Integral fusion of the HtaA CR2 domain from Corynebacterium diphtheriae within EGFP | | Descriptor: | CHLORIDE ION, GLYCEROL, HtaACR2 integral fusion within enhanced green fluorescent protein, ... | | Authors: | Mahoney, B.J, Cascio, D, Clubb, R.T. | | Deposit date: | 2023-04-26 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Development and atomic structure of a new fluorescence-based sensor to probe heme transfer in bacterial pathogens.
J.Inorg.Biochem., 249, 2023
|
|
8U9O
 
 | |
7MPK
 
 | | Crystal structure of TagA with UDP-GlcNAc | | Descriptor: | N-acetylglucosaminyldiphosphoundecaprenol N-acetyl-beta-D-mannosaminyltransferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Martinez, O.E, Cascio, D, Clubb, R.T. | | Deposit date: | 2021-05-04 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.993 Å) | | Cite: | Insight into the molecular basis of substrate recognition by the wall teichoic acid glycosyltransferase TagA.
J.Biol.Chem., 298, 2021
|
|
7M1M
 
 | | Crystal structure of Pseudomonas aeruginosa ClpP1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit | | Authors: | Mawla, G.D, Grant, R.A, Baker, T.A, Sauer, R.T. | | Deposit date: | 2021-03-13 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | ClpP1P2 peptidase activity promotes biofilm formation in Pseudomonas aeruginosa.
Mol.Microbiol., 115, 2021
|
|
7M1L
 
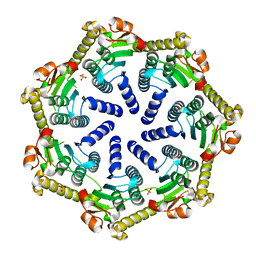 | | Crystal structure of Pseudomonas aeruginosa ClpP2 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, PHOSPHATE ION | | Authors: | Hall, B.M, Grant, R.A, Baker, T.A, Sauer, R.T. | | Deposit date: | 2021-03-13 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ClpP1P2 peptidase activity promotes biofilm formation in Pseudomonas aeruginosa.
Mol.Microbiol., 115, 2021
|
|
7N41
 
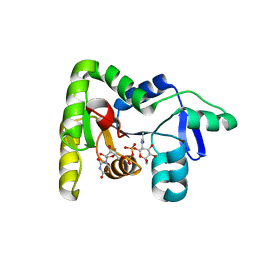 | | Crystal structure of TagA with UDP-ManNAc | | Descriptor: | (2R,3S,4R,5S,6R)-3-acetamido-4,5-dihydroxy-6-(hydroxymethyl)oxan-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]methyl dihydrogen diphosphate (non-preferred name), N-acetylglucosaminyldiphosphoundecaprenol N-acetyl-beta-D-mannosaminyltransferase | | Authors: | Martinez, O.E, Cascio, D, Clubb, R.T. | | Deposit date: | 2021-06-03 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Insight into the molecular basis of substrate recognition by the wall teichoic acid glycosyltransferase TagA.
J.Biol.Chem., 298, 2021
|
|
1IJA
 
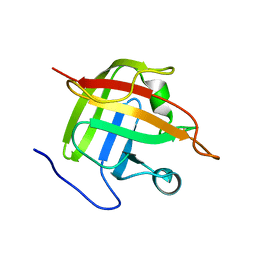 | | Structure of Sortase | | Descriptor: | Sortase | | Authors: | Ilangovan, U, Ton-That, H, Iwahara, J, Schneewind, O, Clubb, R.T. | | Deposit date: | 2001-04-25 | | Release date: | 2001-05-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of sortase, the transpeptidase that anchors proteins to the cell wall of Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1IM1
 
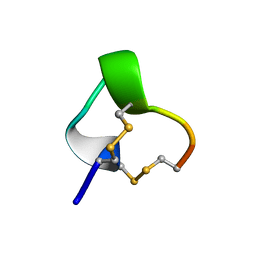 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1, 20 STRUCTURES | | Descriptor: | ALPHA-CONOTOXIN IM1 | | Authors: | Rogers, J.P, Luginbuhl, P, Shen, G.S, Mccabe, R.T, Stevens, R.C, Wemmer, D.E. | | Deposit date: | 1998-11-18 | | Release date: | 1999-06-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of alpha-conotoxin ImI and comparison to other conotoxins specific for neuronal nicotinic acetylcholine receptors.
Biochemistry, 38, 1999
|
|
3L0E
 
 | | X-ray crystal structure of a Potent Liver X Receptor Modulator | | Descriptor: | N-(2-chloro-6-fluorobenzyl)-1-methyl-N-{[3'-(methylsulfonyl)biphenyl-4-yl]methyl}-1H-imidazole-4-sulfonamide, Nuclear receptor coactivator 2, Oxysterols receptor LXR-beta | | Authors: | Gampe Jr, R.T. | | Deposit date: | 2009-12-09 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of tertiary sulfonamides as potent liver X receptor antagonists.
J.Med.Chem., 53, 2010
|
|
1S0T
 
 | | Solution structure of a DNA duplex containing an alpha-anomeric adenosine: insights into substrate recognition by endonuclease IV | | Descriptor: | 5'-D(*Cp*Gp*Tp*Cp*Gp*Tp*Gp*Gp*Ap*C)-3', 5'-D(*Gp*Tp*Cp*Cp*(A3A)p*Cp*Gp*Ap*Cp*G)-3' | | Authors: | Aramini, J.M, Cleaver, S.H, Pon, R.T, Cunningham, R.P, Germann, M.W. | | Deposit date: | 2004-01-04 | | Release date: | 2004-04-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a DNA Duplex Containing an alpha-Anomeric Adenosine: Insights into Substrate Recognition by Endonuclease IV.
J.Mol.Biol., 338, 2004
|
|
1S75
 
 | | SOLUTION STRUCTURE OF A DNA DUPLEX CONTAINING AN ALPHA-ANOMERIC ADENOSINE: INSIGHTS INTO SUBSTRATE RECOGNITION BY ENDONUCLEASE IV | | Descriptor: | 5'-D(*CP*GP*TP*CP*GP*TP*GP*GP*AP*C)-3', 5'-D(*GP*TP*CP*CP*(A3A)P*CP*GP*AP*CP*G)-3' | | Authors: | Aramini, J.M, Cleaver, S.H, Pon, R.T, Cunningham, R.P, Germann, M.W. | | Deposit date: | 2004-01-28 | | Release date: | 2004-04-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a DNA Duplex Containing an alpha-Anomeric Adenosine: Insights into Substrate Recognition by Endonuclease IV.
J.Mol.Biol., 338, 2004
|
|
6WR2
 
 | |
1SJ0
 
 | | Human Estrogen Receptor Alpha Ligand-binding Domain in Complex with the Antagonist Ligand 4-D | | Descriptor: | (2S,3R)-2-(4-(2-(PIPERIDIN-1-YL)ETHOXY)PHENYL)-2,3-DIHYDRO-3-(4-HYDROXYPHENYL)BENZO[B][1,4]OXATHIIN-6-OL, Estrogen receptor | | Authors: | Kim, S, Wu, J.Y, Birzin, E.T, Chan, W, Pai, L.Y, Yang, Y.T, Mosley, R.T, Fitzgerald, P.M, Sharma, N, DiNinno, F, Rohrer, S.P, Schaeffer, J.M, Hammond, M.L. | | Deposit date: | 2004-03-02 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Estrogen Receptor Ligands. II. Discovery of Benzoxathiins as Potent, Selective Estrogen Receptor alpha Modulators.
J.Med.Chem., 47, 2004
|
|
1S74
 
 | | SOLUTION STRUCTURE OF A DNA DUPLEX CONTAINING AN ALPHA-ANOMERIC ADENOSINE: INSIGHTS INTO SUBSTRATE RECOGNITION BY ENDONUCLEASE IV | | Descriptor: | 5'-D(*CP*GP*TP*CP*GP*TP*GP*GP*AP*C)-3', 5'-D(*GP*TP*CP*CP*(A3A)P*CP*GP*AP*CP*G)-3' | | Authors: | Aramini, J.M, Cleaver, S.H, Pon, R.T, Cunningham, R.P, Germann, M.W. | | Deposit date: | 2004-01-28 | | Release date: | 2004-04-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a DNA Duplex Containing an alpha-Anomeric Adenosine: Insights into Substrate Recognition by Endonuclease IV.
J.Mol.Biol., 338, 2004
|
|
6WRF
 
 | | ClpX-ClpP complex bound to GFP-ssrA, recognition complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Fei, X, Sauer, R.T. | | Deposit date: | 2020-04-29 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis of ClpXP recognition and unfolding of ssrA-tagged substrates.
Elife, 9, 2020
|
|
6WSG
 
 | | ClpX-ClpP complex bound to ssrA-tagged GFP, intermediate complex | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, Green fluorescent protein, ... | | Authors: | Fei, X, Sauer, R.T. | | Deposit date: | 2020-04-30 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural basis of ClpXP recognition and unfolding of ssrA-tagged substrates.
Elife, 9, 2020
|
|
2RCE
 
 | |
2RV8
 
 | |
2RUI
 
 | | Solution Structure of the Bacillus anthracis Sortase A-substrate Complex | | Descriptor: | Boc-LPAT*, LPXTG-site transpeptidase family protein | | Authors: | Chan, A.H, Yi, S, Jung, M.E, Clubb, R.T. | | Deposit date: | 2014-06-22 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Bacillus anthracis Sortase A Enzyme Bound to Its Sorting Signal: A FLEXIBLE AMINO-TERMINAL APPENDAGE MODULATES SUBSTRATE ACCESS.
J.Biol.Chem., 290, 2015
|
|
3O1F
 
 | | P1 crystal form of E. coli ClpS at 1.4 A resolution | | Descriptor: | ATP-dependent Clp protease adapter protein clpS | | Authors: | Roman-Hernandez, G, Hou, J.Y, Grant, R.A, Sauer, R.T, Baker, T.A. | | Deposit date: | 2010-07-21 | | Release date: | 2011-07-27 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The ClpS Adaptor Mediates Staged Delivery of N-End Rule Substrates to the AAA+ ClpAP Protease.
Mol.Cell, 43, 2011
|
|
3NPQ
 
 | |
3O2B
 
 | | E. coli ClpS in complex with a Phe N-end rule peptide | | Descriptor: | ATP-dependent Clp protease adaptor protein ClpS, CHLORIDE ION, Phe N-end rule peptide, ... | | Authors: | Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A, de Regt, A. | | Deposit date: | 2010-07-22 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease.
Mol.Cell, 43, 2011
|
|
3O2H
 
 | | E. coli ClpS in complex with a Leu N-end rule peptide | | Descriptor: | ATP-dependent Clp protease adaptor protein ClpS, DNA protection during starvation protein | | Authors: | Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A, de Regt, A. | | Deposit date: | 2010-07-22 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease.
Mol.Cell, 43, 2011
|
|
3O2O
 
 | | Structure of E. coli ClpS ring complex | | Descriptor: | ATP-dependent Clp protease adaptor protein ClpS | | Authors: | Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A, de Regt, A. | | Deposit date: | 2010-07-22 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease.
Mol.Cell, 43, 2011
|
|
