7NO4
 
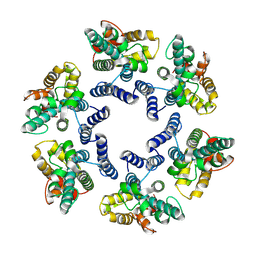 | | Structure of the mature RSV CA lattice: hexamer with 3 adjacent pentamers (C3 symmetric) | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOJ
 
 | | Structure of the mature RSV CA lattice: Group IV, hexamer-hexamer interface, class 3'Beta | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NO3
 
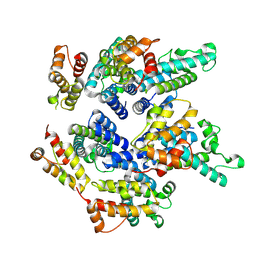 | | Structure of the mature RSV CA lattice: pentamer derived from polyhedral VLPs | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOC
 
 | | Structure of the mature RSV CA lattice: Group III, hexamer-hexamer interface, class 3'3 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOH
 
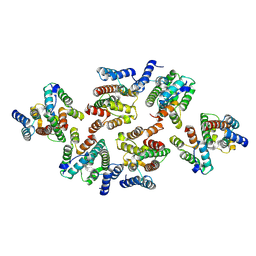 | | Structure of the mature RSV CA lattice: Group III, hexamer-hexamer interface, class 5'5 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOP
 
 | | Structure of the mature RSV CA lattice: Group IV, hexamer-hexamer interface, class 5'Beta | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOA
 
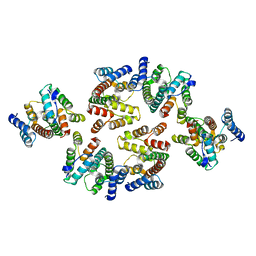 | | Structure of the mature RSV CA lattice: Group II, hexamer-hexamer interface, class 6 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOQ
 
 | | Structure of the mature RSV CA lattice: Group IV, hexamer-hexamer interface, class 5'Gamma | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOF
 
 | | Structure of the mature RSV CA lattice: Group III, hexamer-hexamer interface, class 4'4 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NO6
 
 | | Structure of the mature RSV CA lattice: Group I, pentamer-hexamer interface, class 1 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NON
 
 | | Structure of the mature RSV CA lattice: Group IV, hexamer-hexamer interface, class 4'Gamma | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7MF3
 
 | | Structure of the autoinhibited state of smooth muscle myosin-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Myosin light polypeptide 6, ... | | Authors: | Heissler, S.M, Arora, A.S, Billington, N, Sellers, J.R, Chinthalapudi, K. | | Deposit date: | 2021-04-08 | | Release date: | 2022-01-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the autoinhibited state of myosin-2.
Sci Adv, 7, 2021
|
|
7QGW
 
 | | Sulfonated Calpeptin is a promising drug candidate against SARS-CoV-2 infections | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Calpeptin, ... | | Authors: | Loboda, J, Karnicar, K, Lindic, N, Usenik, A, Lieske, J, Meents, A, Guenther, S, Reinke, P.Y.A, Falke, S, Ewert, W, Turk, D. | | Deposit date: | 2021-12-10 | | Release date: | 2022-12-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
7QKD
 
 | | Crystal structure of human Cathepsin L in complex with covalently bound MG132 | | Descriptor: | ACETATE ION, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 2024
|
|
7QKC
 
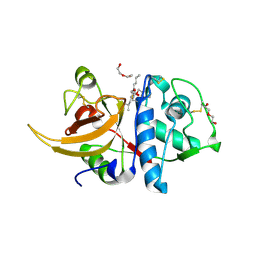 | | Crystal structure of human Cathepsin L after incubation with Sulfo-Calpeptin | | Descriptor: | Calpeptin, Cathepsin L, DI(HYDROXYETHYL)ETHER | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
3MTE
 
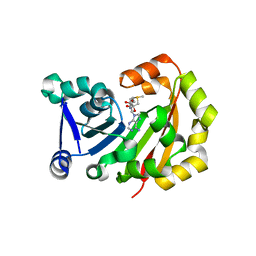 | | Crystal Structure of 16S rRNA Methyltranferase | | Descriptor: | 16S rRNA methylase, S-ADENOSYLMETHIONINE | | Authors: | Macmaster, R.A, Conn, G.L, Zelinskaya, N. | | Deposit date: | 2010-04-30 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Structural insights into the function of aminoglycoside-resistance A1408 16S rRNA methyltransferases from antibiotic-producing and human pathogenic bacteria.
Nucleic Acids Res., 38, 2010
|
|
1XNK
 
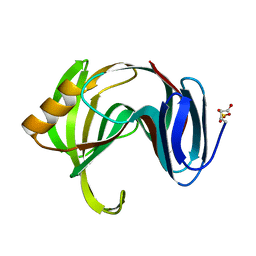 | | Beta-1,4-xylanase from Chaetomium thermophilum complexed with methyl thioxylopentoside | | Descriptor: | 4-thio-beta-D-xylopyranose-(1-4)-4-thio-beta-D-xylopyranose-(1-4)-methyl 4-thio-alpha-D-xylopyranoside, SULFATE ION, endoxylanase 11A | | Authors: | Hakanpaa, J, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2004-10-05 | | Release date: | 2005-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Determination of thioxylo-oligosaccharide binding to family 11 xylanases using electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry and X-ray crystallography
FEBS J., 272, 2005
|
|
1STR
 
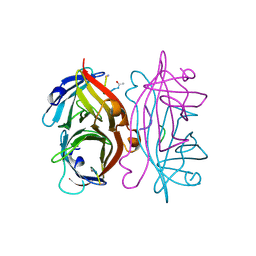 | | STREPTAVIDIN DIMERIZED BY DISULFIDE-BONDED PEPTIDE AC-CHPQNT-NH2 DIMER | | Descriptor: | AC-CHPQNT-NH2, STREPTAVIDIN | | Authors: | Katz, B.A, Cass, R.T, Liu, B, Arze, R, Collins, N. | | Deposit date: | 1995-09-12 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Topochemical catalysis achieved by structure-based ligand design.
J.Biol.Chem., 270, 1995
|
|
1STS
 
 | | STREPTAVIDIN DIMERIZED BY DISULFIDE-BONDED PEPTIDE FCHPQNT-NH2 DIMER | | Descriptor: | FCHPQNT-NH2, STREPTAVIDIN | | Authors: | Katz, B.A, Cass, R.T, Liu, B, Arze, R, Collins, N. | | Deposit date: | 1995-09-12 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Topochemical catalysis achieved by structure-based ligand design.
J.Biol.Chem., 270, 1995
|
|
3Q6T
 
 | | Salivary protein from Lutzomyia longipalpis, Ligand free | | Descriptor: | 43.2 kDa salivary protein, CITRIC ACID | | Authors: | Andersen, J.F, Xu, X, Chang, B.W, Collin, N, Valenzuela, J.G, Ribeiro, J.M. | | Deposit date: | 2011-01-03 | | Release date: | 2011-07-27 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structure and function of a "yellow" protein from saliva of the sand fly Lutzomyia longipalpis that confers protective immunity against Leishmania major infection.
J.Biol.Chem., 286, 2011
|
|
3Q6P
 
 | | Salivary protein from Lutzomyia longipalpis. Selenomethionine derivative | | Descriptor: | 43.2 kDa salivary protein, CITRIC ACID | | Authors: | Andersen, J.F, Xu, X, Chang, B.W, Collin, N, Valenzuela, J.G, Ribeiro, J.M. | | Deposit date: | 2011-01-03 | | Release date: | 2011-07-27 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure and function of a "yellow" protein from saliva of the sand fly Lutzomyia longipalpis that confers protective immunity against Leishmania major infection.
J.Biol.Chem., 286, 2011
|
|
3Q6K
 
 | | Salivary protein from Lutzomyia longipalpis | | Descriptor: | 43.2 kDa salivary protein, CITRIC ACID, SEROTONIN | | Authors: | Andersen, J.F, Xu, X, Chang, B.W, Collin, N, Valenzuela, J.G, Ribeiro, J.M. | | Deposit date: | 2011-01-02 | | Release date: | 2011-07-27 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure and function of a "yellow" protein from saliva of the sand fly Lutzomyia longipalpis that confers protective immunity against Leishmania major infection.
J.Biol.Chem., 286, 2011
|
|
3TTS
 
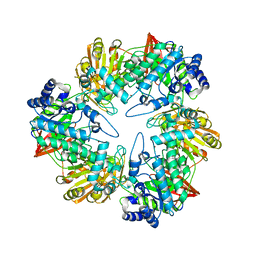 | | Crystal structure of beta-galactosidase from Bacillus circulans sp. alkalophilus | | Descriptor: | Beta-galactosidase, ZINC ION | | Authors: | Maksimainen, M, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2011-09-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural analysis, enzymatic characterization, and catalytic mechanisms of beta-galactosidase from Bacillus circulans sp. alkalophilus.
Febs J., 279, 2012
|
|
3PPS
 
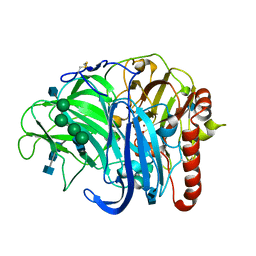 | | Crystal structure of an ascomycete fungal laccase from Thielavia arenaria | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Kallio, J.P, Rouvinen, J, Hakulinen, N. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an ascomycete fungal laccase from Thielavia arenaria--common structural features of asco-laccases.
Febs J., 278, 2011
|
|
3TTY
 
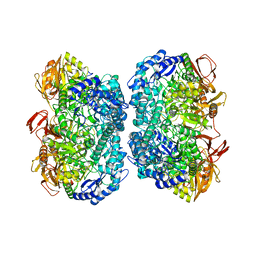 | | Crystal structure of beta-galactosidase from Bacillus circulans sp. alkalophilus in complex with galactose | | Descriptor: | Beta-galactosidase, ZINC ION, alpha-D-galactopyranose | | Authors: | Maksimainen, M, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2011-09-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural analysis, enzymatic characterization, and catalytic mechanisms of beta-galactosidase from Bacillus circulans sp. alkalophilus.
Febs J., 279, 2012
|
|
