1FSE
 
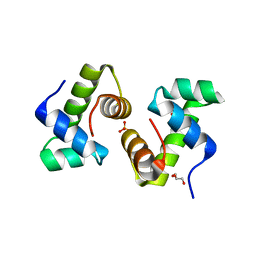 | | CRYSTAL STRUCTURE OF THE BACILLUS SUBTILIS REGULATORY PROTEIN GERE | | Descriptor: | GERE, GLYCEROL, SULFATE ION | | Authors: | Ducros, V.M.-A, Lewis, R.J, Verma, C.S, Dodson, E.J, Leonard, G, Turkenburg, J.P, Murshudov, G.N, Wilkinson, A.J, Brannigan, J.A. | | Deposit date: | 2000-09-08 | | Release date: | 2001-03-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of GerE, the ultimate transcriptional regulator of spore formation in Bacillus subtilis.
J.Mol.Biol., 306, 2001
|
|
1IEN
 
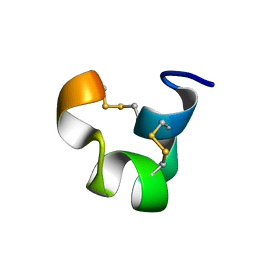 | | SOLUTION STRUCTURE OF TIA | | Descriptor: | PROTEIN TIA | | Authors: | Sharpe, I.A, Gehrmann, J, Loughnan, M.L, Thomas, L, Adams, D.A, Atkins, A, Palant, E, Craik, D.J, Adams, D.J, Alewood, P.F, Lewis, R.J. | | Deposit date: | 2001-04-10 | | Release date: | 2002-04-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Two new classes of conopeptides inhibit the alpha1-adrenoceptor and noradrenaline transporter.
Nat.Neurosci., 4, 2001
|
|
1IEO
 
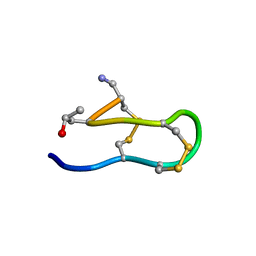 | | SOLUTION STRUCTURE OF MRIB-NH2 | | Descriptor: | PROTEIN MRIB-NH2 | | Authors: | Sharpe, I.A, Gehrmann, J, Loughnan, M.L, Thomas, L, Adams, D.A, Atkins, A, Palant, E, Craik, D.J, Adams, D.J, Alewood, P.F, Lewis, R.J. | | Deposit date: | 2001-04-10 | | Release date: | 2002-04-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Two new classes of conopeptides inhibit the alpha1-adrenoceptor and noradrenaline transporter.
Nat.Neurosci., 4, 2001
|
|
1A39
 
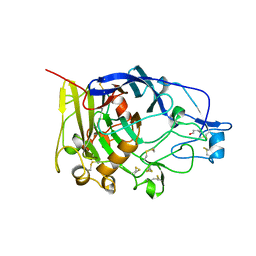 | | HUMICOLA INSOLENS ENDOCELLULASE EGI S37W, P39W DOUBLE-MUTANT | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I | | Authors: | Davies, G.J, Ducros, V, Lewis, R.J, Borchert, T.V, Schulein, M. | | Deposit date: | 1998-01-28 | | Release date: | 1999-03-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Oligosaccharide specificity of a family 7 endoglucanase: insertion of potential sugar-binding subsites.
J.Biotechnol., 57, 1997
|
|
1A0M
 
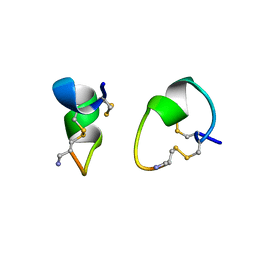 | | 1.1 ANGSTROM CRYSTAL STRUCTURE OF A-CONOTOXIN [TYR15]-EPI | | Descriptor: | ALPHA-CONOTOXIN [TYR15]-EPI | | Authors: | Hu, S.-H, Loughnan, M, Miller, R, Weeks, C.M, Blessing, R.H, Alewood, P.F, Lewis, R.J, Martin, J.L. | | Deposit date: | 1997-12-03 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The 1.1 A resolution crystal structure of [Tyr15]EpI, a novel alpha-conotoxin from Conus episcopatus, solved by direct methods.
Biochemistry, 37, 1998
|
|
1AV3
 
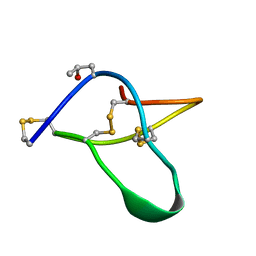 | | POTASSIUM CHANNEL BLOCKER KAPPA CONOTOXIN PVIIA FROM C. PURPURASCENS, NMR, 20 STRUCTURES | | Descriptor: | Kappa-conotoxin PVIIA | | Authors: | Scanlon, M.J, Naranjo, D, Thomas, L, Alewood, P.F, Lewis, R.J, Craik, D.J. | | Deposit date: | 1997-09-24 | | Release date: | 1998-10-14 | | Last modified: | 2020-12-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and proposed binding mechanism of a novel potassium channel toxin kappa-conotoxin PVIIA.
Structure, 5, 1997
|
|
1CNN
 
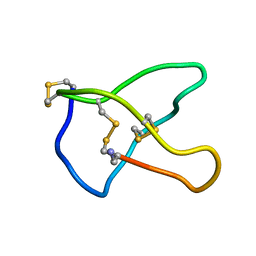 | | OMEGA-CONOTOXIN MVIIC FROM CONUS MAGUS | | Descriptor: | OMEGA-CONOTOXIN MVIIC | | Authors: | Nielsen, K.J, Adams, D, Thomas, L, Bond, T, Alewood, P.F, Craik, D.J, Lewis, R.J. | | Deposit date: | 1999-05-20 | | Release date: | 2000-05-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure-activity relationships of omega-conotoxins MVIIA, MVIIC and 14 loop splice hybrids at N and P/Q-type calcium channels.
J.Mol.Biol., 289, 1999
|
|
1E3A
 
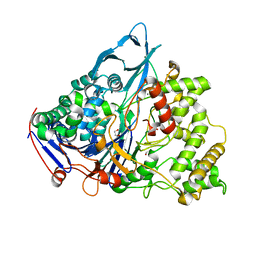 | | A slow processing precursor penicillin acylase from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hewitt, L, Kasche, V, Lummer, K, Lewis, R.J, Murshudov, G.N, Verma, C.S, Dodson, G.G, Wilson, K.S. | | Deposit date: | 2000-06-07 | | Release date: | 2000-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a Slow Processing Precursor Penicillin Acylase from Escherichia Coli Reveals the Linker Peptide Blocking the Active-Site Cleft
J.Mol.Biol., 302, 2000
|
|
1H4Y
 
 | | Structure of the Anti-Sigma Factor Antagonist SpoIIAA in its Unphosphorylated Form | | Descriptor: | ANTI-SIGMA F FACTOR ANTAGONIST | | Authors: | Seavers, P.R, Lewis, R.J, Brannigan, J.A, Verschueren, K.H.G, Murshudov, G.N, Wilkinson, A.J. | | Deposit date: | 2001-05-16 | | Release date: | 2001-07-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structure of the Bacillus Cell Fate Determinant Spoiiaa in Phosphorylated and Unphosphorylated Forms
Structure, 9, 2001
|
|
2A39
 
 | | HUMICOLA INSOLENS ENDOCELLULASE EGI NATIVE STRUCTURE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I | | Authors: | Davies, G.J, Sulzenbacher, G, Mackenzie, L, Withers, S.G, Divne, C, Jones, T.A, Woldike, H.F, Schulein, M. | | Deposit date: | 1998-01-30 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the family 7 endoglucanase I (Cel7B) from Humicola insolens at 2.2 A resolution and identification of the catalytic nucleophile by trapping of the covalent glycosyl-enzyme intermediate.
Biochem.J., 335, 1998
|
|
4MPH
 
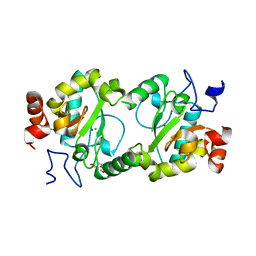 | | Crystal structure of BaLdcB / VanY-like L,D-carboxypeptidase Zinc(II)-bound | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, D-alanyl-D-alanine carboxypeptidase family protein, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Onopriyenko, O, Skarina, T, Shatsman, S, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-12 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.0301 Å) | | Cite: | Structure of the LdcB LD-Carboxypeptidase Reveals the Molecular Basis of Peptidoglycan Recognition.
Structure, 22, 2014
|
|
9BFL
 
 | |
7B7R
 
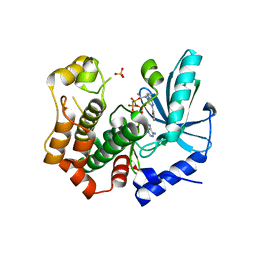 | | MEK1 in complex with compound 4 | | Descriptor: | 2-[5-[ethyl(methyl)amino]imidazo[1,2-a]pyrimidin-7-yl]phenol, Dual specificity mitogen-activated protein kinase kinase 1,Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2020-12-11 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fragment-Based Discovery of Novel Allosteric MEK1 Binders.
Acs Med.Chem.Lett., 12, 2021
|
|
7B3M
 
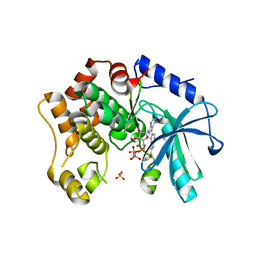 | | MEK1 in complex with compound 6 | | Descriptor: | 1~{H}-indol-2-yl(pyridin-3-yl)methanone, Dual specificity mitogen-activated protein kinase kinase 1,Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2020-12-01 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fragment-Based Discovery of Novel Allosteric MEK1 Binders.
Acs Med.Chem.Lett., 12, 2021
|
|
7B0U
 
 | | Stressosome complex from Listeria innocua | | Descriptor: | RsbR protein, RsbS protein | | Authors: | Miksys, A, Fu, L, Madej, M.G, Ziegler, C. | | Deposit date: | 2020-11-22 | | Release date: | 2021-12-01 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Molecular insights into intra-complex signal transmission during stressosome activation.
Commun Biol, 5, 2022
|
|
7B9L
 
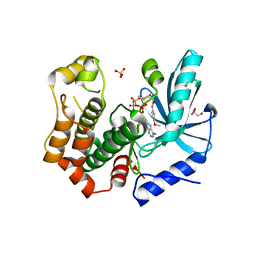 | | MEK1 in complex with compound 23 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1,Dual specificity mitogen-activated protein kinase kinase 1, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fragment-Based Discovery of Novel Allosteric MEK1 Binders.
Acs Med.Chem.Lett., 12, 2021
|
|
7B94
 
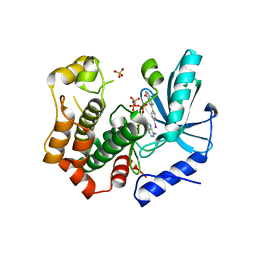 | | MEK1 in complex with compound 6 | | Descriptor: | 2-(4-iodophenyl)-8~{H}-imidazo[1,2-c]pyrimidin-5-one, Dual specificity mitogen-activated protein kinase kinase 1,Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-Based Discovery of Novel Allosteric MEK1 Binders.
Acs Med.Chem.Lett., 12, 2021
|
|
5T90
 
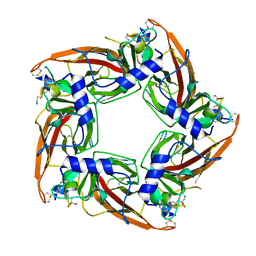 | | Structural mechanisms for alpha-conotoxin selectivity at the human alpha3beta4 nicotinic acetylcholine receptor | | Descriptor: | Acetylcholine-binding protein, LsIA | | Authors: | Abraham, N, Healy, M, Ragnarsson, L, Brust, A, Alewood, P, Lewis, R. | | Deposit date: | 2016-09-08 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanisms for alpha-conotoxin activity at the human alpha 3 beta 4 nicotinic acetylcholine receptor.
Sci Rep, 7, 2017
|
|
6KNO
 
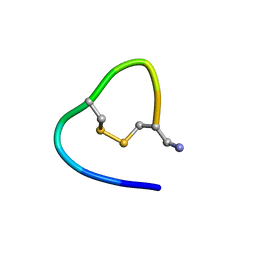 | |
6KMY
 
 | | Structure of single disulfide peptide Czon1107-P5A | | Descriptor: | Czon1107-P5A | | Authors: | Sarma, S.P, Madhan Kumar, M. | | Deposit date: | 2019-08-01 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure and allosteric activity of a single-disulfide conopeptide fromConus zonatusat human alpha 3 beta 4 and alpha 7 nicotinic acetylcholine receptors.
J.Biol.Chem., 295, 2020
|
|
6KN3
 
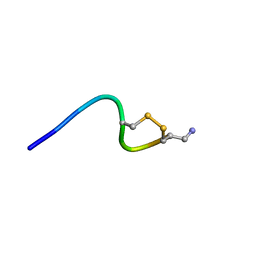 | |
6KNP
 
 | |
6KN2
 
 | |
7Q96
 
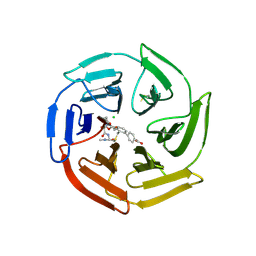 | | Keap1 compound complex | | Descriptor: | 4-[(5S,8R)-5-(dimethylcarbamoyl)-8-[[(2S)-1-ethanoylpyrrolidin-2-yl]carbonylamino]-7,11-bis(oxidanylidene)-10-oxa-3-thia-6-azabicyclo[10.4.0]hexadeca-1(16),12,14-trien-16-yl]benzoic acid, CHLORIDE ION, Kelch-like ECH-associated protein 1 | | Authors: | Johansson, P. | | Deposit date: | 2021-11-12 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.415 Å) | | Cite: | Importance of Binding Site Hydration and Flexibility Revealed When Optimizing a Macrocyclic Inhibitor of the Keap1-Nrf2 Protein-Protein Interaction.
J.Med.Chem., 65, 2022
|
|
7Q6Q
 
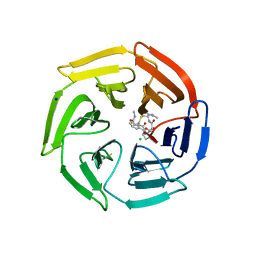 | | Keap1 compound complex | | Descriptor: | (5S,8R)-N,N-dimethyl-8-[[(2S)-1-[4-(methylamino)-4-oxidanylidene-butanoyl]pyrrolidin-2-yl]carbonylamino]-7,11-bis(oxidanylidene)-10-oxa-3-thia-6-azabicyclo[10.4.0]hexadeca-1(12),13,15-triene-5-carboxamide, CHLORIDE ION, Kelch-like ECH-associated protein 1, ... | | Authors: | Johansson, P. | | Deposit date: | 2021-11-09 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Importance of Binding Site Hydration and Flexibility Revealed When Optimizing a Macrocyclic Inhibitor of the Keap1-Nrf2 Protein-Protein Interaction.
J.Med.Chem., 65, 2022
|
|
