4K37
 
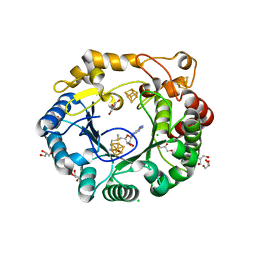 | | Native anSMEcpe with bound AdoMet | | Descriptor: | Anaerobic sulfatase-maturating enzyme, CHLORIDE ION, GLYCEROL, ... | | Authors: | Goldman, P.J, Drennan, C.L. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | X-ray structure of an AdoMet radical activase reveals an anaerobic solution for formylglycine posttranslational modification.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4K36
 
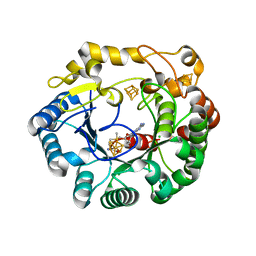 | | His6 tagged anSMEcpe with bound AdoMet | | Descriptor: | Anaerobic sulfatase-maturating enzyme, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Goldman, P.J, Drennan, C.L. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.619 Å) | | Cite: | X-ray structure of an AdoMet radical activase reveals an anaerobic solution for formylglycine posttranslational modification.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4K39
 
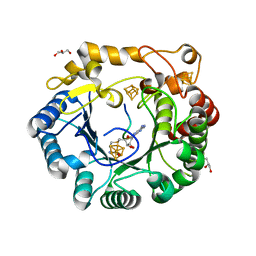 | | Native anSMEcpe with bound AdoMet and Cp18Cys peptide | | Descriptor: | Anaerobic sulfatase-maturating enzyme, CHLORIDE ION, Cp18Cys peptide, ... | | Authors: | Goldman, P.J, Drennan, C.L. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | X-ray structure of an AdoMet radical activase reveals an anaerobic solution for formylglycine posttranslational modification.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4M7S
 
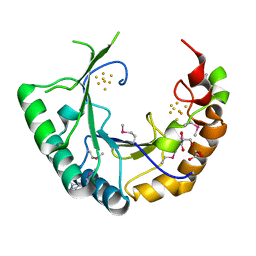 | |
1ZZ6
 
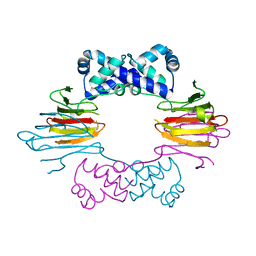 | | Crystal Structure of Apo-HppE | | Descriptor: | Hydroxypropylphosphonic Acid Epoxidase | | Authors: | Higgins, L.J, Yan, F, Liu, P, Liu, H.W, Drennan, C.L. | | Deposit date: | 2005-06-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme
Nature, 437, 2005
|
|
1ZZ9
 
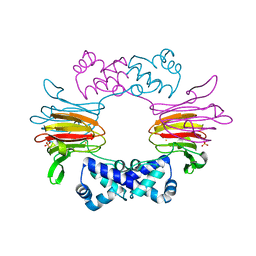 | | Crystal Structure of FeII HppE | | Descriptor: | FE (II) ION, Hydroxypropylphosphonic Acid Epoxidase, SULFATE ION | | Authors: | Higgins, L.J, Yan, F, Liu, P, Liu, H.W, Drennan, C.L. | | Deposit date: | 2005-06-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme
Nature, 437, 2005
|
|
1ZZ7
 
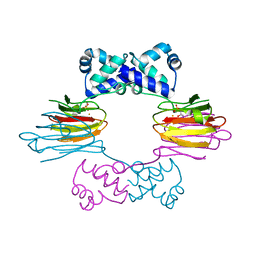 | | Crystal Structure of FeII HppE in Complex with Substrate form 1 | | Descriptor: | (S)-2-HYDROXYPROPYLPHOSPHONIC ACID, FE (II) ION, Hydroxyprophylphosphonic Acid Epoxidase | | Authors: | Higgins, L.J, Yan, F, Liu, P, Liu, H.W, Drennan, C.L. | | Deposit date: | 2005-06-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme
Nature, 437, 2005
|
|
5T6O
 
 | |
5TK7
 
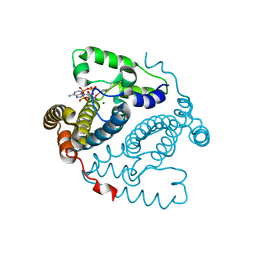 | |
5T81
 
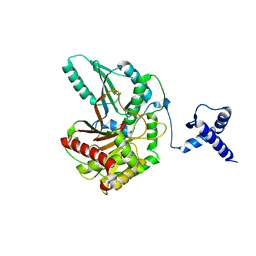 | | Rhombohedral crystal form of the EpoB NRPS cyclization-docking bidomain from Sorangium cellulosum | | Descriptor: | EpoB, GLYCEROL | | Authors: | Dowling, D.P, Kung, Y, Croft, A.K, Taghizadeh, K, Kelly, W.L, Walsh, C.T, Drennan, C.L. | | Deposit date: | 2016-09-06 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Structural elements of an NRPS cyclization domain and its intermodule docking domain.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5T8Y
 
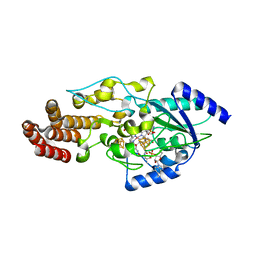 | | Structure of epoxyqueuosine reductase from Bacillus subtilis with the Asp134 catalytic loop swung out of the active site. | | Descriptor: | COBALAMIN, Epoxyqueuosine reductase, IRON/SULFUR CLUSTER, ... | | Authors: | Dowling, D.P, Miles, Z.D, Kohrer, C, Maiocco, S.J, Elliott, S.J, Bandarian, V, Drennan, C.L. | | Deposit date: | 2016-09-08 | | Release date: | 2016-09-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Molecular basis of cobalamin-dependent RNA modification.
Nucleic Acids Res., 44, 2016
|
|
5TKA
 
 | |
5T7Z
 
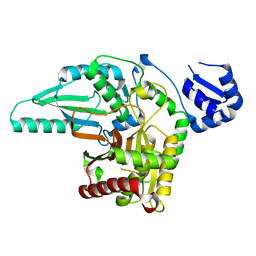 | | Monoclinic crystal form of the EpoB NRPS cyclization-docking bidomain from Sorangium cellulosum | | Descriptor: | EpoB | | Authors: | Dowling, D.P, Kung, Y, Croft, A.K, Taghizadeh, K, Kelly, W.L, Walsh, C.T, Drennan, C.L. | | Deposit date: | 2016-09-06 | | Release date: | 2016-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural elements of an NRPS cyclization domain and its intermodule docking domain.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5TGS
 
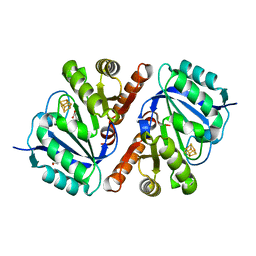 | | Crystal Structure of QueE from Bacillus subtilis with methionine bound | | Descriptor: | 7-carboxy-7-deazaguanine synthase, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Grell, T.A.J, Dowling, D.P, Drennan, C.L. | | Deposit date: | 2016-09-28 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.548 Å) | | Cite: | 7-Carboxy-7-deazaguanine Synthase: A Radical S-Adenosyl-l-methionine Enzyme with Polar Tendencies.
J. Am. Chem. Soc., 139, 2017
|
|
5TK6
 
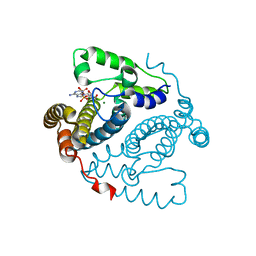 | |
5TH5
 
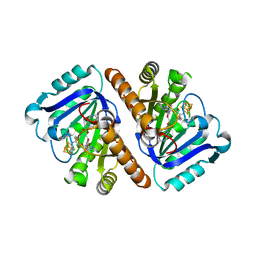 | | Crystal Structure of QueE from Bacillus subtilis with 6-carboxypterin-5'-deoxyadenosyl ester bound | | Descriptor: | 5'-O-(2-amino-4-oxo-1,4-dihydropteridine-6-carbonyl)adenosine, 7-carboxy-7-deazaguanine synthase, IRON/SULFUR CLUSTER, ... | | Authors: | Grell, T.A.J, Dowling, D.P, Drennan, C.L. | | Deposit date: | 2016-09-29 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | 7-Carboxy-7-deazaguanine Synthase: A Radical S-Adenosyl-l-methionine Enzyme with Polar Tendencies.
J. Am. Chem. Soc., 139, 2017
|
|
5TK9
 
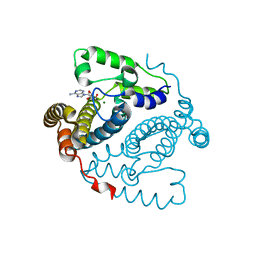 | | Structure of the HD-domain phosphohydrolase OxsA with Oxetanocin-A bound | | Descriptor: | MAGNESIUM ION, OxsA protein, [(2S,3R,4R)-4-(6-amino-9H-purin-9-yl)oxetane-2,3-diyl]dimethanol | | Authors: | Bridwell-Rabb, J, Drennan, C.L. | | Deposit date: | 2016-10-06 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.843 Å) | | Cite: | An HD domain phosphohydrolase active site tailored for oxetanocin-A biosynthesis.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5TK8
 
 | |
2E4G
 
 | | RebH with bound L-Trp | | Descriptor: | TRYPTOPHAN, Tryptophan halogenase | | Authors: | Blasiak, L.C, Drennan, C.L. | | Deposit date: | 2006-12-07 | | Release date: | 2007-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Chlorination by a long-lived intermediate in the mechanism of flavin-dependent halogenases
Biochemistry, 46, 2007
|
|
5UL2
 
 | |
5UL3
 
 | | Structure of Cobalamin-dependent S-adenosylmethionine radical enzyme OxsB with aqua-cobalamin bound | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Bridwell-Rabb, J, Drennan, C.L. | | Deposit date: | 2017-01-24 | | Release date: | 2017-04-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A B12-dependent radical SAM enzyme involved in oxetanocin A biosynthesis.
Nature, 544, 2017
|
|
2E7F
 
 | | 5-methyltetrahydrofolate corrinoid/iron sulfur protein methyltransferase complexed with methyltetrahydrofolate to 2.2 Angsrom resolution | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, 5-methyltetrahydrofolate corrinoid/iron sulfur protein methyltransferase, CALCIUM ION | | Authors: | Doukov, T.I, Drennan, C.L, Hemmi, H, Ragsdale, S.W. | | Deposit date: | 2007-01-09 | | Release date: | 2007-01-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and kinetic evidence for an extended hydrogen-bonding network in catalysis of methyl group transfer. Role of an active site asparagine residue in activation of methyl transfer by methyltransferases.
J.Biol.Chem., 282, 2007
|
|
5UL4
 
 | |
2FCV
 
 | | SyrB2 with Fe(II), bromide, and alpha-ketoglutarate | | Descriptor: | ((2R,3S,4S,5S)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)-5-((2R,3S,4S,5S,6R)-3,4,5-TRIHYDROXY-6-METHOXY-TETRAHYDRO-2H-PYRAN-2-YLOXY)-TETRAHYDROFURAN-2-YL)METHYL NONANOATE, 2-OXOGLUTARIC ACID, BROMIDE ION, ... | | Authors: | Blasiak, L.C, Drennan, C.L. | | Deposit date: | 2005-12-12 | | Release date: | 2006-03-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the non-haem iron halogenase SyrB2 in syringomycin biosynthesis.
Nature, 440, 2006
|
|
2FCT
 
 | | SyrB2 with Fe(II), chloride, and alpha-ketoglutarate | | Descriptor: | ((2R,3S,4S,5S)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)-5-((2R,3S,4S,5S,6R)-3,4,5-TRIHYDROXY-6-METHOXY-TETRAHYDRO-2H-PYRAN-2-YLOXY)-TETRAHYDROFURAN-2-YL)METHYL NONANOATE, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Blasiak, L.C, Drennan, C.L. | | Deposit date: | 2005-12-12 | | Release date: | 2006-03-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the non-haem iron halogenase SyrB2 in syringomycin biosynthesis.
Nature, 440, 2006
|
|
