4RC5
 
 | |
4RT4
 
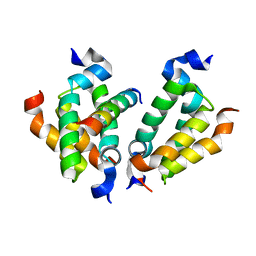 | | Crystal structure of Dpy30 complexed with Bre2 | | Descriptor: | Peptide from COMPASS component BRE2, Protein dpy-30 homolog | | Authors: | Zhang, H.M, Li, M, Chang, W.R. | | Deposit date: | 2014-11-12 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural implications of Dpy30 oligomerization for MLL/SET1 COMPASS H3K4 trimethylation
Protein Cell, 6, 2015
|
|
4RTA
 
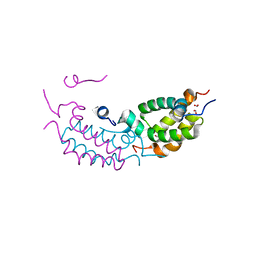 | |
7SCP
 
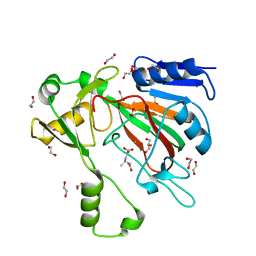 | | The crystal structure of ScoE in complex with intermediate | | Descriptor: | (3R)-3-(oxaloamino)butanoic acid, 1,2-ETHANEDIOL, FE (II) ION, ... | | Authors: | Cha, L, Chen, J, Zhou, J, Chang, W. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Deciphering the Reaction Pathway of Mononuclear Iron Enzyme-Catalyzed N-C Triple Bond Formation in Isocyanide Lipopeptide and Polyketide Biosynthesis
Acs Catalysis, 12, 2022
|
|
7YR5
 
 | |
5W2E
 
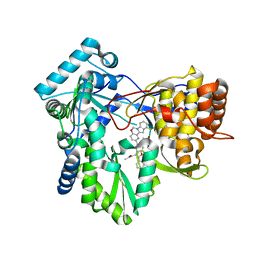 | | HCV NS5B RNA-dependent RNA polymerase in complex with non-nucleoside inhibitor MK-8876 | | Descriptor: | 2-(4-fluorophenyl)-5-(11-fluoro-6H-pyrido[2',3':5,6][1,3]oxazino[3,4-a]indol-2-yl)-N-methyl-6-[methyl(methylsulfonyl)amino]-1-benzofuran-3-carboxamide, Genome polyprotein | | Authors: | Lesburg, C.A, Ummat, A. | | Deposit date: | 2017-06-06 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Development of a New Structural Class of Broadly Acting HCV Non-Nucleoside Inhibitors Leading to the Discovery of MK-8876.
ChemMedChem, 12, 2017
|
|
3PL9
 
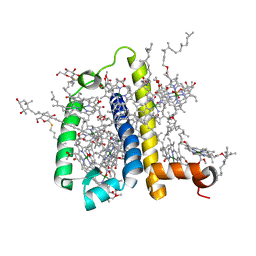 | | Crystal structure of spinach minor light-harvesting complex CP29 at 2.80 angstrom resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X.W, Li, M, Wan, T, Wang, L.F, Jia, C.J, Hou, Z.Q, Zhao, X.L, Zhang, J.P, Chang, W.R. | | Deposit date: | 2010-11-14 | | Release date: | 2011-02-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into energy regulation of light-harvesting complex CP29 from spinach.
Nat.Struct.Mol.Biol., 18, 2011
|
|
6O94
 
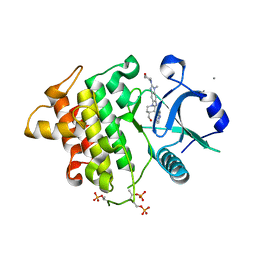 | | Structure of the IRAK4 kinase domain with compound 17 | | Descriptor: | CALCIUM ION, Interleukin-1 receptor-associated kinase 4, N-{5-[4-(hydroxymethyl)piperidin-1-yl]-1-methyl-2-(morpholin-4-yl)-1H-benzimidazol-6-yl}pyrazolo[1,5-a]pyrimidine-3-carboxamide | | Authors: | Yu, C, Drobnick, J, Bryan, M.C, Kiefer, J, Lupardus, P.J. | | Deposit date: | 2019-03-13 | | Release date: | 2019-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Development of Potent and Selective Pyrazolopyrimidine IRAK4 Inhibitors.
J.Med.Chem., 62, 2019
|
|
1WAV
 
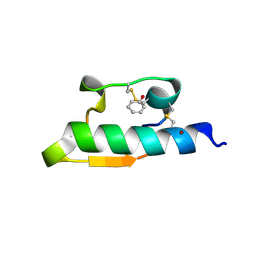 | | CRYSTAL STRUCTURE OF FORM B MONOCLINIC CRYSTAL OF INSULIN | | Descriptor: | INSULIN, PHENOL, ZINC ION | | Authors: | Liang, D.-C, Ding, J.-H, Chang, W.-R, Wan, Z.-L. | | Deposit date: | 1996-02-28 | | Release date: | 1997-02-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular replacement study on form-B monoclinic crystal of insulin.
Sci.China, Ser.C: Life Sci., 39, 1996
|
|
5JYN
 
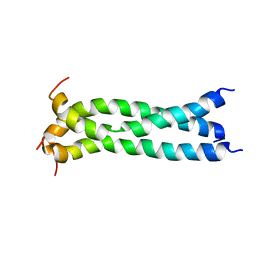 | | Structure of the transmembrane domain of HIV-1 gp41 in bicelle | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Dev, J, Fu, Q, Park, D, Chen, B, Chou, J.J. | | Deposit date: | 2016-05-14 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for membrane anchoring of HIV-1 envelope spike.
Science, 353, 2016
|
|
7MHC
 
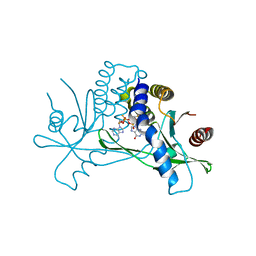 | | Structure of human STING in complex with MK-1454 | | Descriptor: | (2R,5R,7R,8S,10R,12aR,14R,15S,15aR,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-14-(6-amino-9H-purin-9-yl)-15,16-difluoro-2,10-bis(sulfanyl)octahydro-2H,10H,12H-5,8-methano-2lambda~5~,10lambda~5~-furo[3,2-l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Lesburg, C.A. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A kinase-cGAS cascade to synthesize a therapeutic STING activator.
Nature, 603, 2022
|
|
6O9D
 
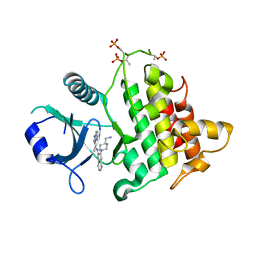 | | Structure of the IRAK4 kinase domain with compound 5 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, N-{7-[4-(aminomethyl)piperidin-1-yl]quinolin-6-yl}pyrazolo[1,5-a]pyrimidine-3-carboxamide | | Authors: | Yu, C, Drobnick, J, Bryan, M.C, Kiefer, J, Lupardus, P.J. | | Deposit date: | 2019-03-13 | | Release date: | 2019-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Development of Potent and Selective Pyrazolopyrimidine IRAK4 Inhibitors.
J.Med.Chem., 62, 2019
|
|
6O8U
 
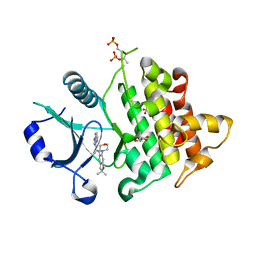 | | Crystal structure of IRAK4 in complex with compound 23 | | Descriptor: | GLYCEROL, Interleukin-1 receptor-associated kinase 4, N-[2,2-dimethyl-6-(morpholin-4-yl)-2,3-dihydro-1-benzofuran-5-yl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, ... | | Authors: | Kiefer, J.R, Yu, C, Drobnick, J, Bryan, M.C, Lupardus, P.J. | | Deposit date: | 2019-03-12 | | Release date: | 2019-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of Potent and Selective Pyrazolopyrimidine IRAK4 Inhibitors.
J.Med.Chem., 62, 2019
|
|
6O95
 
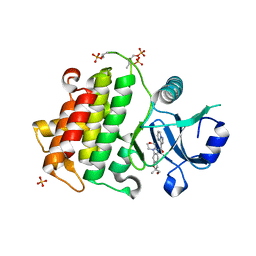 | | Structure of the IRAK4 kinase domain with compound 41 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, N-[(2R)-2-(hydroxymethyl)-2-methyl-6-(morpholin-4-yl)-2,3-dihydro-1-benzofuran-5-yl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, SULFATE ION | | Authors: | Yu, C, Drobnick, J, Bryan, M.C, Kiefer, J, Lupardus, P.J. | | Deposit date: | 2019-03-13 | | Release date: | 2019-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Development of Potent and Selective Pyrazolopyrimidine IRAK4 Inhibitors.
J.Med.Chem., 62, 2019
|
|
3BXV
 
 | |
8GQO
 
 | |
5XNN
 
 | | Structure of M-LHCII and CP24 complexes in the stacked C2S2M2-type PSII-LHCII supercomplex from Pisum sativum | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Su, X.D, Ma, J, Wei, X.P, Cao, P, Zhu, D.J, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2017-05-23 | | Release date: | 2017-09-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex
Science, 357, 2017
|
|
5XNL
 
 | | Structure of stacked C2S2M2-type PSII-LHCII supercomplex from Pisum sativum | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Su, X.D, Ma, J, Wei, X.P, Cao, P, Zhu, D.J, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2017-05-23 | | Release date: | 2017-09-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex
Science, 357, 2017
|
|
5XNM
 
 | | Structure of unstacked C2S2M2-type PSII-LHCII supercomplex from Pisum sativum | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Su, X.D, Ma, J, Wei, X.P, Cao, P, Zhu, D.J, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2017-05-23 | | Release date: | 2017-09-20 | | Last modified: | 2019-05-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex
Science, 357, 2017
|
|
8UPV
 
 | | Structure of SARS-Cov2 3CLPro in complex with Compound 33 | | Descriptor: | 3C-like proteinase nsp5, methyl {(2S)-1-[(1S,3aR,6aS)-1-{[(2R,3S,6R)-6-fluoro-2-hydroxy-1-(methylamino)-1-oxoheptan-3-yl]carbamoyl}hexahydrocyclopenta[c]pyrrol-2(1H)-yl]-3,3-dimethyl-1-oxobutan-2-yl}carbamate | | Authors: | Krishnamurthy, H, Zhuang, N, Qiang, D, Wu, Y, Klein, D.J. | | Deposit date: | 2023-10-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
8UPS
 
 | | Structure of SARS-Cov2 3CLPro in complex with Compound 5 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, PHOSPHATE ION | | Authors: | Wu, Y, Qiang, D, Zhuang, N, Krishnamurthy, H, Klein, D.J. | | Deposit date: | 2023-10-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
8UPW
 
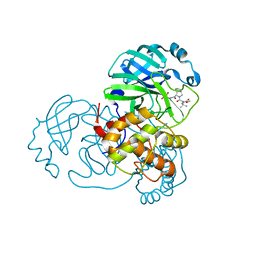 | | Structure of SARS-Cov2 3CLPro in complex with Compound 34 | | Descriptor: | 3C-like proteinase nsp5, methyl {(2S)-1-[(1S,3aR,6aS)-1-{[(2R,3S,6S)-6-fluoro-2-hydroxy-1-(methylamino)-1-oxoheptan-3-yl]carbamoyl}hexahydrocyclopenta[c]pyrrol-2(1H)-yl]-3,3-dimethyl-1-oxobutan-2-yl}carbamate | | Authors: | Krishnamurthy, H, Zhuang, N, Qiang, D, Wu, Y, Klein, D.J. | | Deposit date: | 2023-10-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
8UTE
 
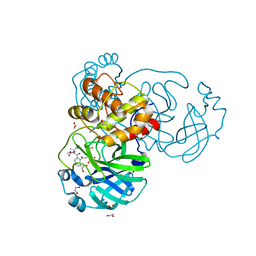 | | Structure of SARS-Cov2 3CLPro in complex with Compound 27 | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, methyl {(2S)-1-[(1S,3aR,6aS)-1-{[(2R,3S)-6,6-difluoro-2-hydroxy-1-(methylamino)-1-oxoheptan-3-yl]carbamoyl}hexahydrocyclopenta[c]pyrrol-2(1H)-yl]-3,3-dimethyl-1-oxobutan-2-yl}carbamate | | Authors: | Krishnamurthy, H, Zhuang, N, Qiang, D, Wu, Y, Klein, D.J. | | Deposit date: | 2023-10-31 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
5XNO
 
 | | Structure of M-LHCII and CP24 complexes in the unstacked C2S2M2-type PSII-LHCII supercomplex from Pisum sativum | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Su, X.D, Ma, J, Wei, X.P, Cao, P, Zhu, D.J, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2017-05-23 | | Release date: | 2017-09-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex
Science, 357, 2017
|
|
3VOP
 
 | | Structure of Vaccinia virus A27 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Protein A27 | | Authors: | Chang, T.H, Ko, T.P, Hsieh, F.L, Wang, A.H.J. | | Deposit date: | 2012-01-31 | | Release date: | 2013-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of vaccinia viral A27 protein reveals a novel structure critical for its function and complex formation with A26 protein.
Plos Pathog., 9, 2013
|
|
