3PQV
 
 | | Cyclase homolog | | Descriptor: | D(-)-TARTARIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Tanaka, N, Smith, P, Shuman, S. | | Deposit date: | 2010-11-27 | | Release date: | 2011-04-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Crystal structure of Rcl1, an essential component of the eukaryal pre-rRNA processosome implicated in 18s rRNA biogenesis.
Rna, 17, 2011
|
|
5V9X
 
 | | Structure of Mycobacterium smegmatis helicase Lhr bound to ssDNA and AMP-PNP | | Descriptor: | ATP-dependent DNA helicase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Ordonez, H, Jacewicz, A, Ferrao, R, Shuman, S. | | Deposit date: | 2017-03-23 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Structure of mycobacterial 3'-to-5' RNA:DNA helicase Lhr bound to a ssDNA tracking strand highlights distinctive features of a novel family of bacterial helicases.
Nucleic Acids Res., 46, 2018
|
|
1RI3
 
 | | Structure and mechanism of mRNA cap (guanine N-7) methyltransferase | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, mRNA CAPPING ENZYME | | Authors: | Fabrega, C, Hausmann, S, Shen, V, Shuman, S, Lima, C.D. | | Deposit date: | 2003-11-16 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of mRNA cap (Guanine-n7) methyltransferase
Mol.Cell, 13, 2004
|
|
1A41
 
 | | TYPE 1-TOPOISOMERASE CATALYTIC FRAGMENT FROM VACCINIA VIRUS | | Descriptor: | SULFATE ION, TOPOISOMERASE I | | Authors: | Cheng, C, Kussie, P, Pavletich, N, Shuman, S. | | Deposit date: | 1998-02-10 | | Release date: | 1999-06-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conservation of structure and mechanism between eukaryotic topoisomerase I and site-specific recombinases.
Cell(Cambridge,Mass.), 92, 1998
|
|
2QZE
 
 | |
2F4Q
 
 | |
2FAQ
 
 | | Crystal Structure of Pseudomonas aeruginosa LigD polymerase domain with ATP and Manganese | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Zhu, H, Nandakumar, J, Aniukwu, J, Wang, L.K, Glickman, M.S, Lima, C.D, Shuman, S. | | Deposit date: | 2005-12-07 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Atomic structure and nonhomologous end-joining function of the polymerase component of bacterial DNA ligase D
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
1RI5
 
 | | Structure and mechanism of mRNA cap (guanine N-7) methyltransferase | | Descriptor: | mRNA CAPPING ENZYME | | Authors: | Fabrega, C, Hausmann, S, Shen, V, Shuman, S, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-11-16 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanism of mRNA cap (Guanine-n7) methyltransferase
Mol.Cell, 13, 2004
|
|
1RI4
 
 | | Structure and mechanism of mRNA cap (guanine N-7) methyltransferase | | Descriptor: | S-ADENOSYLMETHIONINE, mRNA CAPPING ENZYME | | Authors: | Fabrega, C, Hausmann, S, Shen, V, Shuman, S, Lima, C.D. | | Deposit date: | 2003-11-16 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanism of mRNA cap (Guanine-n7) methyltransferase
Mol.Cell, 13, 2004
|
|
1RI2
 
 | | Structure and mechanism of mRNA cap (guanine N-7) methyltransferase | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, mRNA CAPPING ENZYME | | Authors: | Fabrega, C, Hausmann, S, Shen, V, Shuman, S, Lima, C.D. | | Deposit date: | 2003-11-16 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and mechanism of mRNA cap (Guanine-n7) methyltransferase
Mol.Cell, 13, 2004
|
|
8TKZ
 
 | |
8E1H
 
 | | Asp1 kinase in complex with ADP Mg 5-IP7 | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, ADENOSINE-5'-DIPHOSPHATE, Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase, ... | | Authors: | Goldgur, Y, Shuman, S, Benjamin, B. | | Deposit date: | 2022-08-10 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Fission Yeast Inositol Pyrophosphate Kinase Asp1 in Ligand-Free, Substrate-Bound, and Product-Bound States.
Mbio, 13, 2022
|
|
8E1I
 
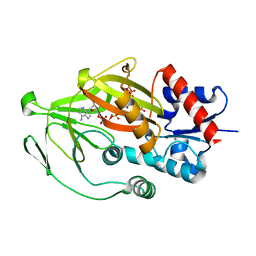 | | Asp1 kinase in complex with ATP Mg 5-IP7 | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, ADENOSINE-5'-TRIPHOSPHATE, Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase, ... | | Authors: | Goldgur, Y, Shuman, S, Benjamin, B. | | Deposit date: | 2022-08-10 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Fission Yeast Inositol Pyrophosphate Kinase Asp1 in Ligand-Free, Substrate-Bound, and Product-Bound States.
Mbio, 13, 2022
|
|
8E1S
 
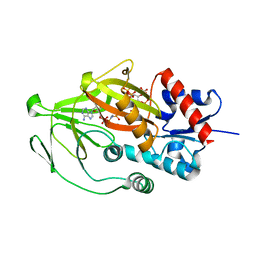 | | Asp1 kinase in complex with ADPNP Mn IP6 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase, MANGANESE (II) ION, ... | | Authors: | Goldgur, Y, Shuman, S, Benjamin, B. | | Deposit date: | 2022-08-11 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structures of Fission Yeast Inositol Pyrophosphate Kinase Asp1 in Ligand-Free, Substrate-Bound, and Product-Bound States.
Mbio, 13, 2022
|
|
8E1T
 
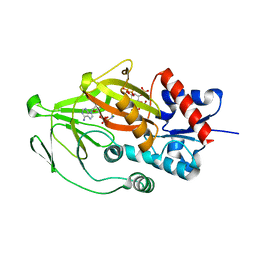 | | Asp1 kinase in complex with ADPNP Mg IP7 | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase, MAGNESIUM ION, ... | | Authors: | Goldgur, Y, Shuman, S, Benjamin, B. | | Deposit date: | 2022-08-11 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structures of Fission Yeast Inositol Pyrophosphate Kinase Asp1 in Ligand-Free, Substrate-Bound, and Product-Bound States.
Mbio, 13, 2022
|
|
8E1J
 
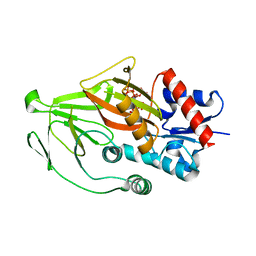 | | Asp1 kinase in complex with 1,5-IP8 | | Descriptor: | (1R,3S,4R,5S,6R)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl bis[trihydrogen (diphosphate)], Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase | | Authors: | Goldgur, Y, Shuman, S, Benjamin, B. | | Deposit date: | 2022-08-10 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of Fission Yeast Inositol Pyrophosphate Kinase Asp1 in Ligand-Free, Substrate-Bound, and Product-Bound States.
Mbio, 13, 2022
|
|
8E1V
 
 | | Asp1 kinase in complex with ADPNP Mg IP6 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase, MAGNESIUM ION, ... | | Authors: | Goldgur, Y, Shuman, S, Benjamin, B. | | Deposit date: | 2022-08-11 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Fission Yeast Inositol Pyrophosphate Kinase Asp1 in Ligand-Free, Substrate-Bound, and Product-Bound States.
Mbio, 13, 2022
|
|
7LFQ
 
 | | Pyrococcus RNA ligase | | Descriptor: | DNA (5'-D(*AP*TP*GP*TP*CP*C)-3'), POTASSIUM ION, RNA-splicing ligase RtcB, ... | | Authors: | Goldgur, Y, Shuman, S, Banerjee, A. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of 3'-PO 4 /5'-OH RNA ligase RtcB in complex with a 5'-OH oligonucleotide.
Rna, 27, 2021
|
|
7MI2
 
 | | Seb1-G476S RNA binding domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Rpb7-binding protein seb1 | | Authors: | Jacewicz, A, Shuman, S. | | Deposit date: | 2021-04-16 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Genetic screen for suppression of transcriptional interference identifies a gain-of-function mutation in Pol2 termination factor Seb1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2IA5
 
 | | T4 polynucleotide kinase/phosphatase with bound sulfate and magnesium. | | Descriptor: | ARSENIC, MAGNESIUM ION, Polynucleotide kinase, ... | | Authors: | Zhu, H, Smith, P.C, Wang, L.K, Lima, C.D, Shuman, S. | | Deposit date: | 2006-09-07 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-function analysis of the 3' phosphatase component of T4 polynucleotide kinase/phosphatase.
Virology, 366, 2007
|
|
7LHL
 
 | |
2LJ6
 
 | |
1P8L
 
 | | New Crystal Structure of Chlorella Virus DNA Ligase-Adenylate | | Descriptor: | ADENOSINE MONOPHOSPHATE, PBCV-1 DNA ligase | | Authors: | Odell, M, Malinina, L, Teplova, M, Shuman, S. | | Deposit date: | 2003-05-07 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Analysis of the DNA Joining Repertoire of Chlorella Virus DNA ligase and a New Crystal Structure of the Ligase-Adenylate Intermediate
Nucleic Acids Res., 31, 2003
|
|
1P16
 
 | | Structure of an mRNA capping enzyme bound to the phosphorylated carboxyl-terminal domain of RNA polymerase II | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Fabrega, C, Shen, V, Shuman, S, Lima, C.D. | | Deposit date: | 2003-04-11 | | Release date: | 2003-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of an mRNA capping enzyme bound to the phosphorylated carboxy-terminal domain of RNA polymerase II.
Mol.Cell, 11, 2003
|
|
4GP6
 
 | | Polynucleotide kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Metallophosphoesterase | | Authors: | Wang, L.K, Das, U, Smith, P, Shuman, S. | | Deposit date: | 2012-08-20 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanism of the polynucleotide kinase component of the bacterial Pnkp-Hen1 RNA repair system.
Rna, 18, 2012
|
|
