8GHY
 
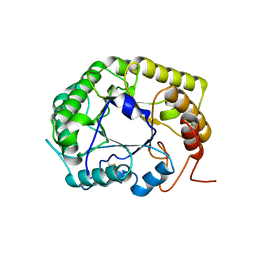 | | Crystal Structure of the E154D mutant CelD Cellulase from the Anaerobic Fungus Piromyces finnis in the complex with cellotriose. | | Descriptor: | Cellulase CelD, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Dementieve, A, Kim, Y, Jedrzejczak, R, Michalska, K, Joachimiak, A. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and enzymatic characterization of CelD endoglucanase from the anaerobic fungus Piromyces finnis.
Appl.Microbiol.Biotechnol., 107, 2023
|
|
8G62
 
 | | Papain-Like Protease of SARS CoV-2 in complex with remodilin NCGC 390004 | | Descriptor: | 3-methoxy-5-(1-methylpiperidin-4-yl)-N-[4-(pyrrolidine-1-sulfonyl)phenyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Jedrzejczak, R, Luci, D, Kales, S, Simeonov, A, Rai, G, Drayman, N, Tay, S, Oakes, S, Rosner, M, Chen, B, Dulin, N, Solway, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-02-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Papain-Like Protease of SARS CoV-2 in complex with remodilin NCGC 390004
To Be Published
|
|
8UW6
 
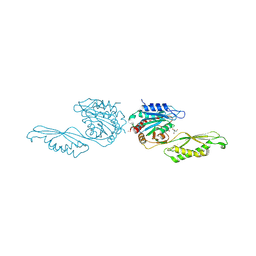 | | Acetylornithine deacetylase from Escherichia coli, di-zinc form. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Acetylornithine deacetylase, ... | | Authors: | Osipiuk, J, Endres, M, Kelley, E, Becker, D.P, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-11-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | N alpha-acetyl-L-ornithine deacetylase from Escherichia coli and a ninhydrin-based assay to enable inhibitor identification.
Front Chem, 12, 2024
|
|
8TTP
 
 | | Crystal structure of class C beta-lactamase from Escherichia coli in complex with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-08-14 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure of class C beta-lactamase from Escherichia coli in complex with avibactam
to be published
|
|
8U00
 
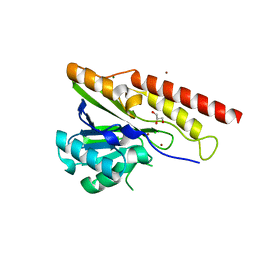 | | Crystal structure of metallo-beta-lactamase superfamily protein from Caulobacter vibrioides | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-08-28 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of metallo-beta-lactamase superfamily protein from Caulobacter vibrioides
To Be Published
|
|
8U12
 
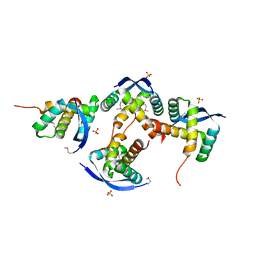 | |
4M9D
 
 | | The Crystal structure of an adenylosuccinate synthetase from Bacillus anthracis str. Ames Ancestor in complex with AMP. | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Adenylosuccinate synthetase, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-14 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | The Crystal structure of an adenylosuccinate synthetase from Bacillus anthracis str. Ames Ancestor in complex with AMP.
To be Published
|
|
4LQB
 
 | | Crystal structure of uncharacterized protein Kfla3161 | | Descriptor: | CITRIC ACID, GLYCEROL, Uncharacterized protein | | Authors: | Chang, C, Chhor, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-07-17 | | Release date: | 2013-07-31 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of uncharacterized protein Kfla3161
To be Published
|
|
4M88
 
 | |
4LZH
 
 | | L,D-transpeptidase from Klebsiella pneumoniae | | Descriptor: | L,D-transpeptidase | | Authors: | Osipiuk, J, Hatzos-Skintges, C, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-07-31 | | Release date: | 2013-08-21 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | L,D-transpeptidase from Klebsiella pneumoniae.
To be Published
|
|
4M0M
 
 | | The crystal structure of a functionally unknown protein lpg2422 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | PHOSPHATE ION, Putative uncharacterized protein, TETRAETHYLENE GLYCOL | | Authors: | Tan, K, Li, H, Clancy, S, Shuman, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-08-01 | | Release date: | 2013-08-21 | | Method: | X-RAY DIFFRACTION (2.192 Å) | | Cite: | The crystal structure of a functionally unknown protein lpg2422 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1
To be Published
|
|
4MAA
 
 | |
4MI1
 
 | | Crystal structure of the double mutant (S112A, H303A) of B.anthracis mycrocine immunity protein (MccF) with aspartyl sulfamoyl adenylates | | Descriptor: | 5'-O-(L-alpha-aspartylsulfamoyl)adenosine, Microcin immunity protein MccF, SULFATE ION | | Authors: | Nocek, B, Severinov, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-30 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the double mutant (S112A, H303A) of B.anthracis mycrocine immunity protein (MccF) with aspartyl sulfamoyl adenylates
TO BE PUBLISHED
|
|
4M0C
 
 | | The crystal structure of a FMN-dependent NADH-azoreductase from Bacillus anthracis str. Ames Ancestor in complex with FMN. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase 1, GLYCEROL, ... | | Authors: | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-01 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.073 Å) | | Cite: | The crystal structure of a FMN-dependent NADH-azoreductase from Bacillus anthracis str. Ames Ancestor in complex with FMN.
To be Published
|
|
4M0G
 
 | | The crystal structure of an adenylosuccinate synthetase from Bacillus anthracis str. Ames Ancestor. | | Descriptor: | Adenylosuccinate synthetase, CHLORIDE ION | | Authors: | Tan, K, Zhou, M, Zhang, R, Kwon, K, Anderson, W.F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-01 | | Release date: | 2013-08-14 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | The crystal structure of an adenylosuccinate synthetase from Bacillus anthracis str. Ames Ancestor.
To be Published
|
|
4MLZ
 
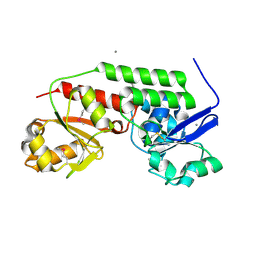 | | Crystal structure of periplasmic binding protein from Jonesia denitrificans | | Descriptor: | CALCIUM ION, POTASSIUM ION, Periplasmic binding protein | | Authors: | Chang, C, Chhor, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-06 | | Release date: | 2013-09-18 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of periplasmic binding protein from Jonesia denitrificans
To be Published
|
|
4MJX
 
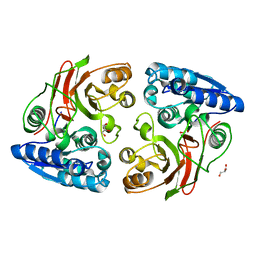 | |
4MLC
 
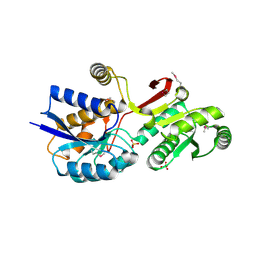 | | ABC Transporter Substrate-Binding Protein fromDesulfitobacterium hafniense | | Descriptor: | CALCIUM ION, Extracellular ligand-binding receptor, SULFATE ION | | Authors: | Kim, Y, Chhor, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-06 | | Release date: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | ABC Transporter Substrate-Binding Protein fromDesulfitobacterium hafniense
To be Published
|
|
4MJM
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Short Internal Deletion of CBS Domain from Bacillus anthracis str. Ames | | Descriptor: | 1,2-ETHANEDIOL, Inosine-5'-monophosphate dehydrogenase | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-03 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2544 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Short Internal Deletion of CBS Domain from Bacillus anthracis str. Ames
To be Published, 2013
|
|
4MK6
 
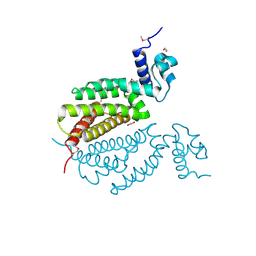 | | Crystal Structure of Probable Dihydroxyacetone Kinase Regulator DHSK_reg from Listeria monocytogenes EGD-e | | Descriptor: | 1,2-ETHANEDIOL, Probable Dihydroxyacetone Kinase Regulator DHSK_reg | | Authors: | Kim, Y, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-04 | | Release date: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Probable Dihydroxyacetone Kinase Regulator DHSK_reg from Listeria monocytogenes EGD-e
To be Published
|
|
4MO9
 
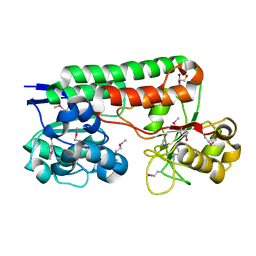 | | Crystal Structure of TroA-like Periplasmic Binding Protein FepB from Veillonella parvula | | Descriptor: | GLYCEROL, Periplasmic binding protein, trimethylamine oxide | | Authors: | Kim, Y, Wu, R, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-11 | | Release date: | 2013-09-25 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | Crystal Structure of TroA-like Periplasmic Binding Protein FepB from Veillonella parvula
To be Published
|
|
4MQB
 
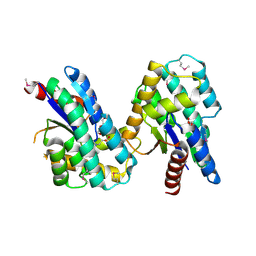 | | Crystal structure of thymidylate kinase from Staphylococcus aureus in complex with 2-(N-morpholino)ethanesulfonic acid | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, TETRAETHYLENE GLYCOL, Thymidylate kinase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2013-09-16 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of thymidylate kinase from Staphylococcus aureus in complex with 2-(N-morpholino)ethanesulfonic acid
To be Published
|
|
4ML9
 
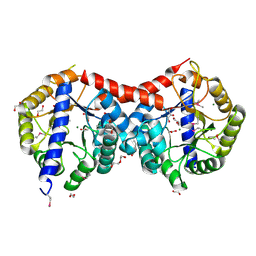 | | Crystal Structure of Uncharacterized TIM Barrel Protein with the Conserved Phosphate Binding Site fromSebaldella termitidis | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kim, Y, Holowicki, J, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-06 | | Release date: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | Crystal Structure of Uncharacterized TIM Barrel Protein with the Conserved Phosphate Binding Site fromSebaldella termitidis
To be Published
|
|
4MNU
 
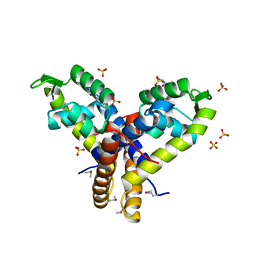 | | Crystal Structure of Uncharacterized SlyA-like Transcription Regulator from Listeria monocytogenes | | Descriptor: | GLYCEROL, SULFATE ION, SlyA-like transcription regulator | | Authors: | Kim, Y, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-11 | | Release date: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal Structure of Uncharacterized SlyA-like Transcription Regulator from
Listeria monocytogenes
To be Published
|
|
4MY3
 
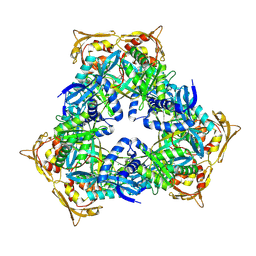 | | Crystal Structure of GCN5-related N-acetyltransferase from Kribbella flavida | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GCN5-related N-acetyltransferase | | Authors: | Kim, Y, Mack, J, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-27 | | Release date: | 2013-10-09 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal Structure of GCN5-related N-acetyltransferase from Kribbella flavida
To be Published
|
|
