3OPK
 
 | | Crystal structure of divalent-cation tolerance protein CutA from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 | | Descriptor: | ACETATE ION, Divalent-cation tolerance protein cutA, MAGNESIUM ION, ... | | Authors: | Nocek, B, Mulligan, R, Papazisi, L, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-01 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of divalent-cation tolerance protein CutA from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2
TO BE PUBLISHED
|
|
3Q10
 
 | | Pantoate-beta-alanine ligase from Yersinia pestis | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Osipiuk, J, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-16 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Pantoate-beta-alanine ligase from Yersinia pestis.
To be Published
|
|
3PLX
 
 | | The crystal structure of aspartate alpha-decarboxylase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | ACETATE ION, Aspartate 1-decarboxylase, DI(HYDROXYETHYL)ETHER | | Authors: | Tan, K, Gu, M, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-11-15 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | The crystal structure of aspartate alpha-decarboxylase from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
3Q0I
 
 | | Methionyl-tRNA formyltransferase from Vibrio cholerae | | Descriptor: | GLYCEROL, Methionyl-tRNA formyltransferase, SULFATE ION | | Authors: | Osipiuk, J, Gu, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-15 | | Release date: | 2010-12-29 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Methionyl-tRNA formyltransferase from Vibrio cholerae.
To be Published
|
|
4MY8
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor Q21 | | Descriptor: | (2S)-2-(naphthalen-1-yloxy)-N-[2-(pyridin-4-yl)-1,3-benzoxazol-5-yl]propanamide, 1,2-ETHANEDIOL, ACETIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Kavitha, M, Cuny, G, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-27 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2924 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor Q21
To be Published, 2013
|
|
4MZ8
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with an Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound C91 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-29 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5004 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound C91
To be Published
|
|
4E16
 
 | | Precorrin-4 C(11)-methyltransferase from Clostridium difficile | | Descriptor: | precorrin-4 C(11)-methyltransferase | | Authors: | Osipiuk, J, Nocek, B, Makowska-Grzyska, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-05 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Precorrin-4 C(11)-methyltransferase from Clostridium difficile
To be Published
|
|
4MZ1
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound P12 | | Descriptor: | 1-(4-bromophenyl)-3-{2-[3-(prop-1-en-2-yl)phenyl]propan-2-yl}urea, ACETIC ACID, INOSINIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-28 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3991 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound P12
To be Published, 2013
|
|
4QMK
 
 | | Crystal structure of type III effector protein ExoU (exoU) | | Descriptor: | BETA-MERCAPTOETHANOL, Type III secretion system effector protein ExoU | | Authors: | Halavaty, A.S, Tyson, G.H, Zhang, A, Hauser, A.R, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-16 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Novel Phosphatidylinositol 4,5-Bisphosphate Binding Domain Mediates Plasma Membrane Localization of ExoU and Other Patatin-like Phospholipases.
J.Biol.Chem., 290, 2015
|
|
3Q12
 
 | | Pantoate-beta-alanine ligase from Yersinia pestis in complex with pantoate. | | Descriptor: | CHLORIDE ION, PANTOATE, Pantoate--beta-alanine ligase | | Authors: | Osipiuk, J, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-16 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Pantoate-beta-alanine ligase from Yersinia pestis.
To be Published
|
|
4FFY
 
 | | Crystal structure of DENV1-E111 single chain variable fragment bound to DENV-1 DIII, strain 16007. | | Descriptor: | CHLORIDE ION, DENV1-E111 single chain variable fragment (heavy chain), DENV1-E111 single chain variable fragment (light chain), ... | | Authors: | Austin, S.K, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-06-01 | | Release date: | 2012-06-20 | | Last modified: | 2012-10-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of Differential Neutralization of DENV-1 Genotypes by an Antibody that Recognizes a Cryptic Epitope.
Plos Pathog., 8, 2012
|
|
3UPB
 
 | | 1.5 Angstrom Resolution Crystal Structure of Transaldolase from Francisella tularensis in Covalent Complex with Arabinose-5-Phosphate | | Descriptor: | ARABINOSE-5-PHOSPHATE, DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, ... | | Authors: | Light, S.H, Minasov, G, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-17 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Arabinose 5-phosphate covalently inhibits transaldolase.
J.Struct.Funct.Genom., 15, 2014
|
|
3GJZ
 
 | | Crystal structure of microcin immunity protein MccF from Bacillus anthracis str. Ames | | Descriptor: | Microcin immunity protein MccF | | Authors: | Nocek, B, Zhou, M, Kwon, K, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-03-09 | | Release date: | 2009-04-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Functional Characterization of Microcin C Resistance Peptidase MccF from Bacillus anthracis.
J.Mol.Biol., 420, 2012
|
|
4FFE
 
 | | The structure of cowpox virus CPXV018 (OMCP) | | Descriptor: | CPXV018 protein | | Authors: | Lazear, E, Peterson, L.W, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-06-01 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Cowpox Virus-Encoded NKG2D Ligand OMCP.
J.Virol., 87, 2013
|
|
7KPP
 
 | | Structure of the E102A mutant of a GNAT superfamily PA3944 acetyltransferase | | Descriptor: | 1,2-ETHANEDIOL, Acetyltransferase PA3944, COENZYME A, ... | | Authors: | Czub, M.P, Porebski, P.J, Majorek, K.A, Cymborowski, M, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-12 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Gcn5-Related N- Acetyltransferases (GNATs) With a Catalytic Serine Residue Can Play Ping-Pong Too.
Front Mol Biosci, 8, 2021
|
|
7KYE
 
 | | Structure of a GNAT superfamily PA3944 acetyltransferase in complex with CHES | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Acetyltransferase PA3944, ... | | Authors: | Czub, M.P, Porebski, P.J, Cymborowski, M, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of a GNAT superfamily PA3944 acetyltransferase in complex with CHES
To Be Published
|
|
7KYU
 
 | | The crystal structure of SARS-CoV-2 Main Protease with the formation of Cys145-1H-indole-5-carboxylate | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1H-indole-5-carbonyl)oxy]-1H-benzotriazole, 3C-like proteinase | | Authors: | Tan, K, Maltseva, N.I, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-08 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The crystal structure of SARS-CoV-2 Main Protease with the formation of Cys145-1H-indole-5-carboxylate
To Be Published
|
|
7KZW
 
 | | Crystal structure of FTT_1639c from Francisella tularensis str. tularensis SCHU S4 | | Descriptor: | CHLORIDE ION, FTT_1639c | | Authors: | Stogios, P.J, Skarina, T, Osipiuk, J, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-10 | | Release date: | 2020-12-30 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal structure of FTT_1639c from Francisella tularensis str. tularensis SCHU S4
To Be Published
|
|
3M6L
 
 | | Crystal structure of transketolase in complex with thiamine diphosphate, ribose-5-phosphate and calcium ion | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Nocek, B, Makowska-Grzyska, M, Maltseva, N, Joachimiak, A, Anderson, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-15 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal structure of transketolase in complex with thiamine diphosphate, ribose-5-phosphate and calcium ion
To be Published
|
|
7KPS
 
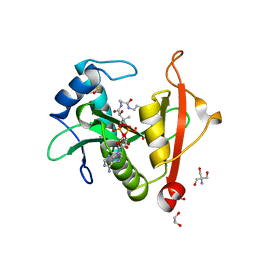 | | Structure of a GNAT superfamily PA3944 acetyltransferase in complex with AcCoA | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYL COENZYME *A, ... | | Authors: | Czub, M.P, Porebski, P.J, Cymborowski, M, Reidl, C.T, Becker, D.P, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-12 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Gcn5-Related N- Acetyltransferases (GNATs) With a Catalytic Serine Residue Can Play Ping-Pong Too.
Front Mol Biosci, 8, 2021
|
|
4ONW
 
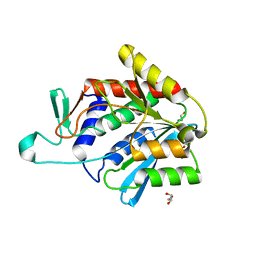 | | Crystal structure of the catalytic domain of DapE protein from V.cholerea | | Descriptor: | 1,2-ETHANEDIOL, 1,4-BUTANEDIOL, ACETATE ION, ... | | Authors: | Nocek, B, Makowska-Grzyska, M, Jedrzejczak, R, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-29 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Dimerization Domain in DapE Enzymes Is required for Catalysis.
Plos One, 9, 2014
|
|
3MUE
 
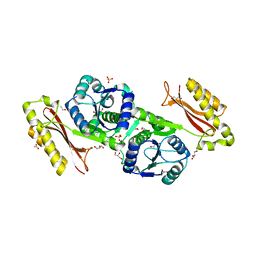 | | Crystal Structure of Pantoate-beta-Alanine Ligase from Salmonella typhimurium | | Descriptor: | ACETIC ACID, ETHANOL, GLYCEROL, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-03 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystal Structure of Pantoate-beta-Alanine Ligase from Salmonella typhimurium
To be Published
|
|
4OP4
 
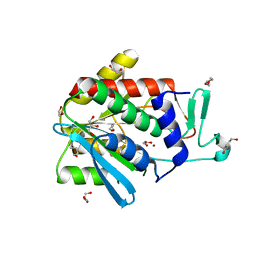 | | Crystal structure of the catalytic domain of DapE protein from V.cholerea in the Zn bound form | | Descriptor: | 1,2-ETHANEDIOL, 1,4-BUTANEDIOL, GLYCEROL, ... | | Authors: | Nocek, B, Makowska-Grzyska, M, Jedrzejczak, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-02-04 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | The Dimerization Domain in DapE Enzymes Is required for Catalysis.
Plos One, 9, 2014
|
|
3ERP
 
 | | Structure of IDP01002, a putative oxidoreductase from and essential gene of Salmonella typhimurium | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Singer, A.U, Minasov, G, Evdokimova, E, Brunzelle, J.S, Kudritska, M, Edwards, A.M, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-10-02 | | Release date: | 2008-11-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and biochemical studies of novel aldo-keto reductases for the biocatalytic conversion of 3-hydroxybutanal to 1,3-butanediol.
Appl.Environ.Microbiol., 2017
|
|
3NZT
 
 | | 2.0 Angstrom Crystal structure of Glutamate--Cysteine Ligase (gshA) ftom Francisella tularensis in Complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glutamate--cysteine ligase, SULFATE ION | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-16 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 Angstrom Crystal structure of Glutamate--Cysteine Ligase (gshA) ftom Francisella tularensis in Complex with AMP.
To be Published
|
|
