3T62
 
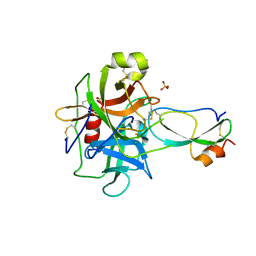 | | Crystal structure of recombinant Kunitz Type serine protease Inhibitor-1 from the Caribbean Sea anemone Stichodactyla helianthus in complex with bovine chymotrypsin | | Descriptor: | Chymotrypsinogen A, Kunitz-type proteinase inhibitor SHPI-1, SULFATE ION | | Authors: | Garcia-Fernandez, R, Dominguez, R, Oberthuer, D, Pons, T, Gonzalez-Gonzalez, Y, Chavez, M.A, Betzel, C, Redecke, L. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into chymotrypsin inhibition by the Kunitz-type inhibitor-1 from the marine invertebrate Stichodactyla helianthus
To be Published
|
|
6EZF
 
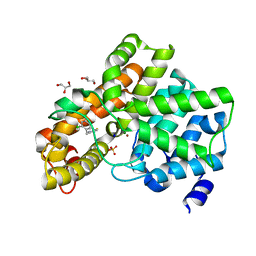 | | PDE2 in complex with molecule 5 | | Descriptor: | 6-[(2,4-dichlorophenyl)methyl]pyridazine-3-thiol, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Tresadern, G, Perez-Benito, L, Keraenen, H, van Vlijmen, H. | | Deposit date: | 2017-11-15 | | Release date: | 2018-04-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Predicting Binding Free Energies of PDE2 Inhibitors. The Difficulties of Protein Conformation.
Sci Rep, 8, 2018
|
|
4C33
 
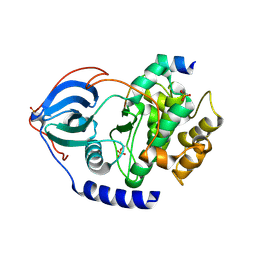 | | PKA-S6K1 Chimera Apo | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA | | Authors: | Couty, S, Westwood, I.M, Kalusa, A, Cano, C, Travers, J, Boxall, K, Chow, C.L, Burns, S, Schmitt, J, Pickard, L, Barillari, C, McAndrew, P.C, Clarke, P.A, Linardopoulos, S, Griffin, R.J, Aherne, G.W, Raynaud, F.I, Workman, P, Jones, K, van Montfort, R.L.M. | | Deposit date: | 2013-08-21 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The discovery of potent ribosomal S6 kinase inhibitors by high-throughput screening and structure-guided drug design.
Oncotarget, 4, 2013
|
|
1N5G
 
 | |
4BW2
 
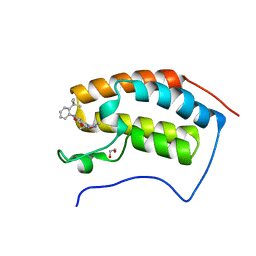 | | The first bromodomain of human BRD4 in complex with 3,5 dimethylisoxaxole ligand | | Descriptor: | 1,2-ETHANEDIOL, 4-((2-(TERT-BUTYL)PHENYL)AMINO)-7-(3,5-dimethylisoxazol-4-yl)-1,8-naphthyridine-3-carboxylic acid, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Chung, C, Mirguet, O, Lamotte, Y, Bamborough, P, Delannee, D, Bouillot, A, Gellibert, F, Krysa, G, Lewis, A, Witherington, J, Huet, P, Dudit, Y, Trottet, L, Nicodeme, E. | | Deposit date: | 2013-06-29 | | Release date: | 2013-09-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Naphthyridines as Novel Bet Family Bromodomain Inhibitors.
Chemmedchem, 9, 2014
|
|
6EZQ
 
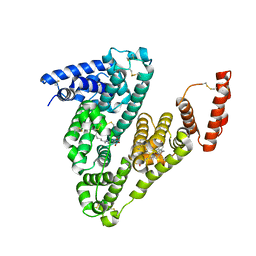 | | human Serum Albumin complexed with NBD-C12 fatty acid | | Descriptor: | 12-[(4-nitro-2,1,3-benzoxadiazol-7-yl)amino]dodecanoic acid, Serum albumin | | Authors: | Wenskowsky, L, Liesum, A, Schreuder, H.A. | | Deposit date: | 2017-11-16 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Identification and Characterization of a Single High-Affinity Fatty Acid Binding Site in Human Serum Albumin.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Y4Z
 
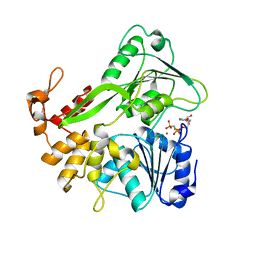 | | Crystal structure of the Zika virus NS3 helicase complex with AMPPNP | | Descriptor: | MANGANESE (II) ION, NS3 helicase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Li, L. | | Deposit date: | 2017-08-06 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural view of the helicase reveals that Zika virus uses a conserved mechanism for unwinding RNA
Acta Crystallogr.,Sect.F, 74, 2018
|
|
8BW3
 
 | | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative | | Descriptor: | (2S)-N-(cyclopropylmethyl)-2-methyl-4-(1-methyl-1H-pyrrole-2-carbonyl)piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassell-Hart, S, Bradshaw, W.J, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2022-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative
To Be Published
|
|
6F1E
 
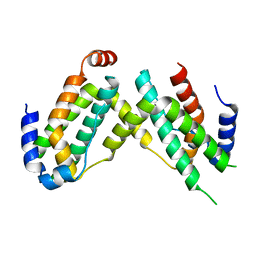 | | Crystal structure of olive flounder [Paralichthys olivaceus] interferon gamma at 2.3 Angstrom resolution | | Descriptor: | Interferon gamma | | Authors: | Kolenko, P, Kolarova, L, Zahradnik, J, Schneider, B. | | Deposit date: | 2017-11-21 | | Release date: | 2018-05-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.296 Å) | | Cite: | Interferons type II and their receptors R1 and R2 in fish species: Evolution, structure, and function.
Fish Shellfish Immunol., 79, 2018
|
|
6F1K
 
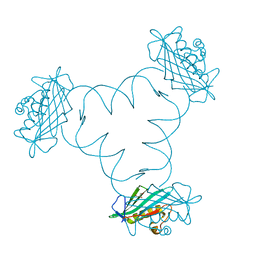 | | Structure of ARTD2/PARP2 WGR domain bound to double strand DNA without 5'phosphate | | Descriptor: | CHLORIDE ION, DNA (5'-D(*GP*CP*CP*TP*AP*GP*CP*TP*AP*CP*GP*TP*AP*GP*CP*TP*AP*GP*GP*C)-3'), GLYCEROL, ... | | Authors: | Obaji, E, Haikarainen, T, Lehtio, L. | | Deposit date: | 2017-11-22 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for DNA break recognition by ARTD2/PARP2.
Nucleic Acids Res., 46, 2018
|
|
3T77
 
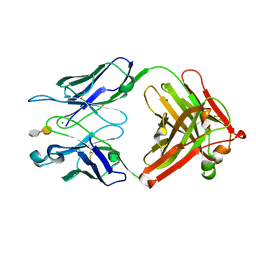 | | S25-2- A(2-4)KDO disaccharide complex | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, MAGNESIUM ION, S25-2 FAB (IGG1K) heavy chain, ... | | Authors: | Nguyen, H.P, Seto, N.O, Mackenzie, C.R, Brade, L, Kosma, P, Brade, H, Evans, S.V. | | Deposit date: | 2011-07-29 | | Release date: | 2011-08-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.739 Å) | | Cite: | Germline antibody recognition of distinct carbohydrate epitopes.
Nat.Struct.Biol., 10, 2003
|
|
4BPU
 
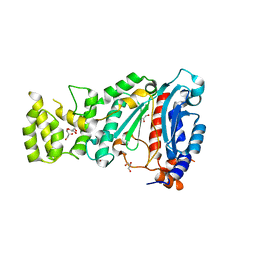 | | Crystal structure of human primase in heterodimeric form, comprising PriS and truncated PriL lacking the C-terminal Fe-S domain. | | Descriptor: | DNA PRIMASE LARGE SUBUNIT, DNA PRIMASE SMALL SUBUNIT, GLYCEROL, ... | | Authors: | Kilkenny, M.L, Perera, R.L, Pellegrini, L. | | Deposit date: | 2013-05-28 | | Release date: | 2013-09-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of Human Primase Reveal Design of Nucleotide Elongation Site and Mode of Pol Alpha Tethering
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6F1U
 
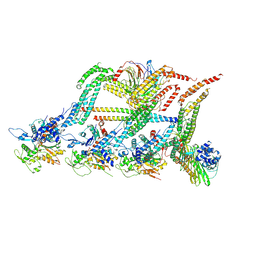 | | N terminal region of dynein tail domains in complex with dynactin filament and BICDR-1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARP1 actin related protein 1 homolog A, BICD family-like cargo adapter 1, ... | | Authors: | Urnavicius, L, Lau, C.K, Elshenawy, M.M, Morales-Rios, E, Motz, C, Yildiz, A, Carter, A.P. | | Deposit date: | 2017-11-23 | | Release date: | 2018-01-17 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM shows how dynactin recruits two dyneins for faster movement.
Nature, 554, 2018
|
|
6F38
 
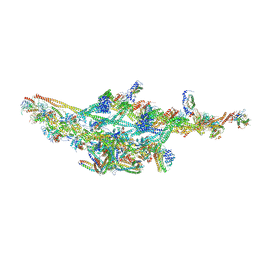 | | Cryo-EM structure of two dynein tail domains bound to dynactin and HOOK3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ARP1 actin related protein 1 homolog A, ... | | Authors: | Lau, C.K, Urnavicius, L, Elshenawy, M.M, Morales-Rios, E, Motz, C, Yildiz, A, Carter, A.P. | | Deposit date: | 2017-11-28 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Cryo-EM shows how dynactin recruits two dyneins for faster movement.
Nature, 554, 2018
|
|
8BW2
 
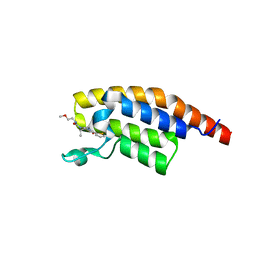 | | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative | | Descriptor: | (2R)-N-(2-methoxyethyl)-2-methyl-4-thiophen-2-ylcarbonyl-piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassell-Hart, S, Bradshaw, W.J, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2022-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative
To Be Published
|
|
5YAV
 
 | | Fragment-based Drug Discovery of inhibitors to block PDEdelta-RAS protein-protein interaction | | Descriptor: | 1-phenyl-5-propan-2-ylsulfanyl-1,2,3,4-tetrazole, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Bing, X, Yanlian, L, Danyan, C. | | Deposit date: | 2017-09-01 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | Fragment-based Drug Discovery of inhibitors to block PDEdelta-RAS protein-protein interaction
To Be Published
|
|
5M5X
 
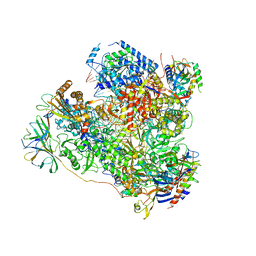 | | RNA Polymerase I elongation complex 1 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Tafur, L, Sadian, Y, Hoffmann, N.A, Jakobi, A.J, Wetzel, R, Hagen, W.J.H, Sachse, C, Muller, C.W. | | Deposit date: | 2016-10-23 | | Release date: | 2016-12-21 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular Structures of Transcribing RNA Polymerase I.
Mol. Cell, 64, 2016
|
|
4BQJ
 
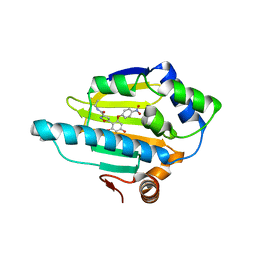 | | structure of HSP90 with an inhibitor bound | | Descriptor: | 5-[2,4-dihydroxy-6-(4-nitrophenoxy)phenyl]-N-ethyl-1,2-oxazole-3-carboxamide, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Casale, E, Brasca, M.G, Mantegani, S, Amboldi, N, Bindi, S, Caronni, D, Ceccarelli, W, Colombo, N, DePonti, A, Donati, D, Ermoli, A, Fachin, G, Felder, E.R, Ferguson, R.D, Fiorelli, C, Guanci, M, Isacchi, A, Pesenti, E, Polucci, P, Riceputi, L, Sola, F, Visco, C, Zuccotto, F, Fogliatto, G. | | Deposit date: | 2013-05-30 | | Release date: | 2013-10-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Nms-E973 as Novel, Selective and Potent Inhibitor of Heat Shock Protein 90 (Hsp90).
Bioorg.Med.Chem., 21, 2013
|
|
4BU3
 
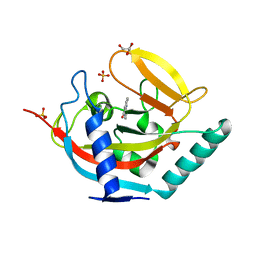 | | Crystal structure of human tankyrase 2 in complex with 2-phenyl-3,4- dihydroquinazolin-4-one | | Descriptor: | 2-phenyl-3H-quinazolin-4-one, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Para-Substituted 2-Phenyl-3,4-Dihydroquinazolin-4-Ones as Potent and Selective Tankyrase Inhibitors.
Chemmedchem, 8, 2013
|
|
1OB4
 
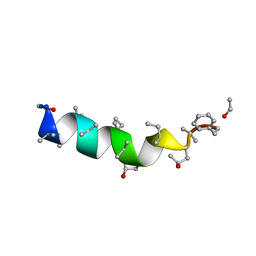 | | Cephaibol A | | Descriptor: | CEPHAIBOL A, ETHANOL | | Authors: | Bunkoczi, G, Schiell, M, Vertesy, L, Sheldrick, G.M. | | Deposit date: | 2003-01-24 | | Release date: | 2003-12-11 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Crystal Structures of Cephaibols
J.Pept.Sci., 9, 2003
|
|
3TBM
 
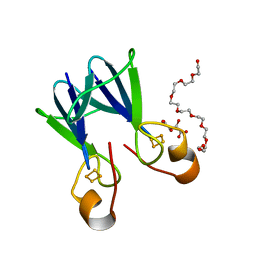 | | Crystal structure of a type 4 CDGSH iron-sulfur protein. | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, L(+)-TARTARIC ACID, NONAETHYLENE GLYCOL, ... | | Authors: | Lin, J, Zhang, L, Ye, K. | | Deposit date: | 2011-08-07 | | Release date: | 2011-10-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Structure and Molecular Evolution of CDGSH Iron-Sulfur Domains.
Plos One, 6, 2011
|
|
5YKG
 
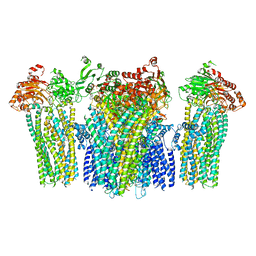 | |
1OB6
 
 | | Cephaibol B | | Descriptor: | ACETATE ION, CEPHAIBOL B, ETHANOL | | Authors: | Bunkoczi, G, Schiell, M, Vertesy, L, Sheldrick, G.M. | | Deposit date: | 2003-01-24 | | Release date: | 2003-12-11 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Crystal Structures of Cephaibols
J.Pept.Sci., 9, 2003
|
|
5MAM
 
 | | Human insulin in complex with serotonin | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Brzozowski, A.M, Turkenburg, J.P, Jiracek, J, Zakova, L. | | Deposit date: | 2016-11-03 | | Release date: | 2017-04-05 | | Last modified: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Computational and structural evidence for neurotransmitter-mediated modulation of the oligomeric states of human insulin in storage granules.
J. Biol. Chem., 292, 2017
|
|
5YM0
 
 | | The crystal structure of DHAD | | Descriptor: | Dihydroxy-acid dehydratase, chloroplastic, SULFATE ION | | Authors: | Zang, X, Huang, W.X, Cheng, R, Wu, L, Zhou, J.H, Tang, Y, Yan, Y. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | The crystal structure of DHAD
To Be Published
|
|
