3RIZ
 
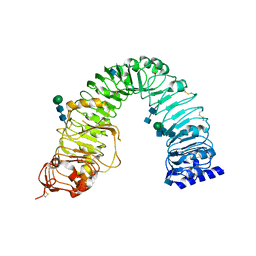 | | Crystal structure of the plant steroid receptor BRI1 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein BRASSINOSTEROID INSENSITIVE 1, ... | | Authors: | Hothorn, M. | | Deposit date: | 2011-04-14 | | Release date: | 2011-06-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.523 Å) | | Cite: | Structural basis of steroid hormone perception by the receptor kinase BRI1.
Nature, 474, 2011
|
|
2OOK
 
 | |
2OOC
 
 | |
2F58
 
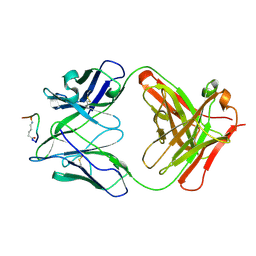 | | IGG1 FAB FRAGMENT (58.2) COMPLEX WITH 12-RESIDUE CYCLIC PEPTIDE (INCLUDING RESIDUES 315-324 OF HIV-1 GP120) (MN ISOLATE) | | Descriptor: | 1-IMINO-5-PENTANONE, PROTEIN (HIV-1 GP120), PROTEIN (IGG1 FAB 58.2 ANTIBODY (HEAVY CHAIN)), ... | | Authors: | Stanfield, R.L, Cabezas, E, Satterthwait, A.C, Stura, E.A, Profy, A.T, Wilson, I.A. | | Deposit date: | 1998-10-23 | | Release date: | 1999-02-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dual conformations for the HIV-1 gp120 V3 loop in complexes with different neutralizing fabs.
Structure Fold.Des., 7, 1999
|
|
1F58
 
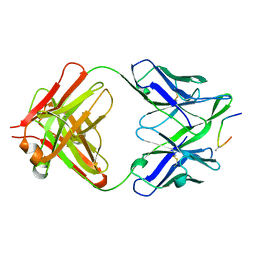 | | IGG1 FAB FRAGMENT (58.2) COMPLEX WITH 24-RESIDUE PEPTIDE (RESIDUES 308-333 OF HIV-1 GP120 (MN ISOLATE) WITH ALA TO AIB SUBSTITUTION AT POSITION 323 | | Descriptor: | Envelope glycoprotein gp120, PROTEIN (IGG1 ANTIBODY 58.2 (HEAVY CHAIN)), PROTEIN (IGG1 ANTIBODY 58.2 (LIGHT CHAIN)) | | Authors: | Stanfield, R.L, Cabezas, E, Satterthwait, A.C, Stura, E.A, Profy, A.T, Wilson, I.A. | | Deposit date: | 1998-10-21 | | Release date: | 1999-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dual conformations for the HIV-1 gp120 V3 loop in complexes with different neutralizing fabs.
Structure Fold.Des., 7, 1999
|
|
1HTM
 
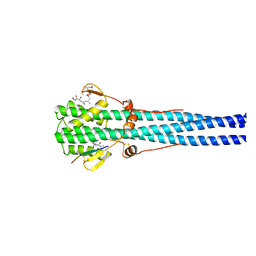 | | STRUCTURE OF INFLUENZA HAEMAGGLUTININ AT THE PH OF MEMBRANE FUSION | | Descriptor: | HEMAGGLUTININ HA1 CHAIN, HEMAGGLUTININ HA2 CHAIN | | Authors: | Bullough, P.A, Hughson, F.M, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1994-11-02 | | Release date: | 1995-02-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of influenza haemagglutinin at the pH of membrane fusion.
Nature, 371, 1994
|
|
3BOS
 
 | |
3CM1
 
 | |
3D00
 
 | |
3DEE
 
 | |
1PSH
 
 | |
3E0F
 
 | |
3DUE
 
 | |
3B77
 
 | |
3BY7
 
 | |
3BYQ
 
 | |
3DCX
 
 | |
3DL1
 
 | |
3CGH
 
 | |
3EQX
 
 | |
3ETN
 
 | |
3G23
 
 | |
3G0T
 
 | |
3FKQ
 
 | |
3HN7
 
 | |
