1VQB
 
 | | GENE V PROTEIN (SINGLE-STRANDED DNA BINDING PROTEIN) | | Descriptor: | GENE V PROTEIN | | Authors: | Skinner, M.M, Zhang, H, Leschnitzer, D.H, Guan, Y, Bellamy, H, Sweet, R.M, Gray, C.W, Konings, R.N.H, Wang, A.H.-J, Terwilliger, T.C. | | Deposit date: | 1996-08-14 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the gene V protein of bacteriophage f1 determined by multiwavelength x-ray diffraction on the selenomethionyl protein.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1VQG
 
 | |
1VQI
 
 | | GENE V PROTEIN MUTANT WITH ILE 47 REPLACED BY VAL 47 (I47V) | | Descriptor: | GENE V PROTEIN | | Authors: | Zhang, H, Skinner, M.M, Sandberg, W.S, Wang, A.H.-J, Terwilliger, T.C. | | Deposit date: | 1996-08-14 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Context dependence of mutational effects in a protein: the crystal structures of the V35I, I47V and V35I/I47V gene V protein core mutants.
J.Mol.Biol., 259, 1996
|
|
1VQF
 
 | | GENE V PROTEIN MUTANT WITH VAL 35 REPLACED BY ILE 35 AND ILE 47 REPLACED BY VAL 47 (V35I, I47V) | | Descriptor: | GENE V PROTEIN | | Authors: | Zhang, H, Skinner, M.M, Sandberg, W.S, Wang, A.H.-J, Terwilliger, T.C. | | Deposit date: | 1996-08-14 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Context dependence of mutational effects in a protein: the crystal structures of the V35I, I47V and V35I/I47V gene V protein core mutants.
J.Mol.Biol., 259, 1996
|
|
1VQJ
 
 | | GENE V PROTEIN MUTANT WITH VAL 35 REPLACED BY ILE 35 (V35I) | | Descriptor: | GENE V PROTEIN | | Authors: | Zhang, H, Skinner, M.M, Sandberg, W.S, Wang, A.H.-J, Terwilliger, T.C. | | Deposit date: | 1996-08-14 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Context dependence of mutational effects in a protein: the crystal structures of the V35I, I47V and V35I/I47V gene V protein core mutants.
J.Mol.Biol., 259, 1996
|
|
1C2T
 
 | | NEW INSIGHTS INTO INHIBITOR DESIGN FROM THE CRYSTAL STRUCTURE AND NMR STUDIES OF E. COLI GAR TRANSFORMYLASE IN COMPLEX WITH BETA-GAR AND 10-FORMYL-5,8,10-TRIDEAZAFOLIC ACID. | | Descriptor: | 10-FORMYL-5,8,10-TRIDEAZAFOLIC ACID, GLYCINAMIDE RIBONUCLEOTIDE, GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Authors: | Greasley, S.E, Yamashita, M.M, Cai, H, Benkovic, S.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 1999-07-26 | | Release date: | 2000-01-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New insights into inhibitor design from the crystal structure and NMR studies of Escherichia coli GAR transformylase in complex with beta-GAR and 10-formyl-5,8,10-trideazafolic acid.
Biochemistry, 38, 1999
|
|
2HES
 
 | | Cytosolic Iron-sulphur Assembly Protein- 1 | | Descriptor: | CALCIUM ION, Ydr267cp | | Authors: | Srinivasan, V, Michel, H, Lill, R, Daili, J.A.N, Pierik, A.J. | | Deposit date: | 2006-06-22 | | Release date: | 2007-07-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Yeast WD40 Domain Protein Cia1, a Component Acting Late in Iron-Sulfur Protein Biogenesis.
Structure, 15, 2007
|
|
2ICI
 
 | |
2NN7
 
 | | Structure of inhibitor binding to Carbonic Anhydrase I | | Descriptor: | Carbonic anhydrase 1, DIMETHYL SULFOXIDE, ETHYL 3-[4-(AMINOSULFONYL)PHENYL]PROPANOATE, ... | | Authors: | Christianson, D.W, Jude, K.M. | | Deposit date: | 2006-10-23 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Analysis of Charge Discrimination in the Binding of Inhibitors to Human Carbonic Anhydrases I and II.
J.Am.Chem.Soc., 129, 2007
|
|
2NNG
 
 | | Structure of inhibitor binding to Carbonic Anhydrase II | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONAMIDE, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Christianson, D.W, Jude, K.M. | | Deposit date: | 2006-10-24 | | Release date: | 2007-05-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Analysis of Charge Discrimination in the Binding of Inhibitors to Human Carbonic Anhydrases I and II.
J.Am.Chem.Soc., 129, 2007
|
|
2NNS
 
 | |
2NN1
 
 | | Structure of inhibitor binding to Carbonic Anhydrase I | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[4-(AMINOSULFONYL)PHENYL]PROPANOIC ACID, Carbonic anhydrase 1, ... | | Authors: | Christianson, D.W, Jude, K.M. | | Deposit date: | 2006-10-23 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Analysis of Charge Discrimination in the Binding of Inhibitors to Human Carbonic Anhydrases I and II
J.Am.Chem.Soc., 129, 2007
|
|
2NNO
 
 | | Structure of inhibitor binding to Carbonic Anhydrase II | | Descriptor: | 3-[4-(AMINOSULFONYL)PHENYL]PROPANOIC ACID, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Christianson, D.W, Jude, K.M. | | Deposit date: | 2006-10-24 | | Release date: | 2007-05-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Structural Analysis of Charge Discrimination in the Binding of Inhibitors to Human Carbonic Anhydrases I and II.
J.Am.Chem.Soc., 129, 2007
|
|
2NMX
 
 | | Structure of inhibitor binding to Carbonic Anhydrase I | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Carbonic anhydrase 1, N-{2-[4-(AMINOSULFONYL)PHENYL]ETHYL}ACETAMIDE, ... | | Authors: | Christianson, D.W, Jude, K.M. | | Deposit date: | 2006-10-23 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Analysis of Charge Discrimination in the Binding of Inhibitors to Human Carbonic Anhydrases I and II
J.Am.Chem.Soc., 129, 2007
|
|
2NNV
 
 | | Structure of inhibitor binding to Carbonic Anhydrase II | | Descriptor: | Carbonic anhydrase 2, ETHYL 3-[4-(AMINOSULFONYL)PHENYL]PROPANOATE, GLYCEROL, ... | | Authors: | Christianson, D.W, Jude, K.M. | | Deposit date: | 2006-10-24 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural Analysis of Charge Discrimination in the Binding of Inhibitors to Human Carbonic Anhydrases I and II.
J.Am.Chem.Soc., 129, 2007
|
|
1LPF
 
 | |
1XQI
 
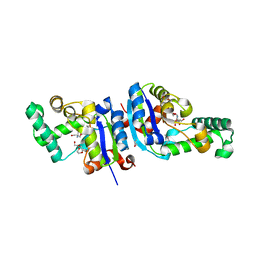 | | Crystal Structure Analysis of an NDP kinase from Pyrobaculum aerophilum | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Nucleoside diphosphate kinase, TRIETHYLENE GLYCOL | | Authors: | Pedelacq, J.D, Waldo, G.S, Cabantous, S, Liong, E.C, Berendzen, J, Terwilliger, T.C. | | Deposit date: | 2004-10-12 | | Release date: | 2005-09-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional features of an NDP kinase from the hyperthermophile crenarchaeon Pyrobaculum aerophilum
Protein Sci., 14, 2005
|
|
1VQD
 
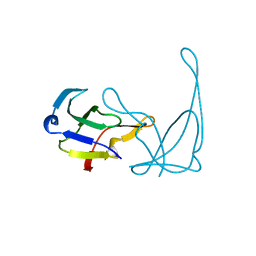 | |
1VQC
 
 | |
1VQE
 
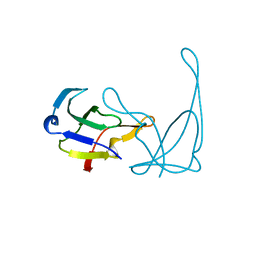 | |
1PPD
 
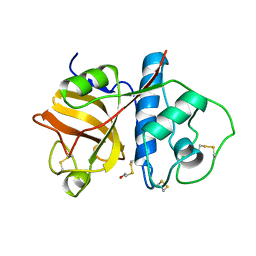 | |
1YQC
 
 | | Crystal Structure of Ureidoglycolate Hydrolase (AllA) from Escherichia coli O157:H7 | | Descriptor: | GLYOXYLIC ACID, Ureidoglycolate hydrolase | | Authors: | Raymond, S, Tocilj, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2005-02-01 | | Release date: | 2005-10-18 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.709 Å) | | Cite: | Crystal structure of ureidoglycolate hydrolase (AllA) from Escherichia coli O157:H7
Proteins, 61, 2005
|
|
1JPZ
 
 | | Crystal structure of a complex of the heme domain of P450BM-3 with N-Palmitoylglycine | | Descriptor: | BIFUNCTIONAL P-450:NADPH-P450 REDUCTASE, N-PALMITOYLGLYCINE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Haines, D.C, Tomchick, D.R, Machius, M, Peterson, J.A. | | Deposit date: | 2001-08-03 | | Release date: | 2001-11-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Pivotal role of water in the mechanism of P450BM-3.
Biochemistry, 40, 2001
|
|
1SEN
 
 | | Endoplasmic reticulum protein Rp19 O95881 | | Descriptor: | CHLORIDE ION, PLATINUM (II) ION, Thioredoxin-like protein p19 | | Authors: | Liu, Z.-J, Chen, L, Tempel, W, Shah, A, Lee, D, Dailey, T.A, Mayer, M.R, Rose, J.P, Richardson, D.C, Richardson, J.S, Dailey, H.A, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-02-17 | | Release date: | 2004-07-13 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Endoplasmic reticulum protein Rp19
To be Published
|
|
1F63
 
 | | CRYSTAL STRUCTURE OF DEOXY SPERM WHALE MYOGLOBIN MUTANT Y(B10)Q(E7)R(E10) | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Brunori, M, Cutruzzola, F, Savino, C, Travaglini-Allocatelli, C, Vallone, B, Gibson, Q.H. | | Deposit date: | 2000-06-20 | | Release date: | 2000-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural dynamics of ligand diffusion in the protein matrix: A study on a new myoglobin mutant Y(B10) Q(E7) R(E10).
Biophys.J., 76, 1999
|
|
