6G47
 
 | | Crystal Structure of Human Adenovirus 52 Short Fiber Knob in Complex with alpha-(2,8)-Trisialic Acid (DP3) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Liaci, A.M, Stehle, T. | | Deposit date: | 2018-03-26 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Polysialic acid is a cellular receptor for human adenovirus 52.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7ZH8
 
 | | DYRK1a in Complex with a Bromo-Triazolo-Pyridine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 6-bromanyl-3H-[1,2,3]triazolo[4,5-b]pyridine, CHLORIDE ION, ... | | Authors: | Dammann, M, Stahlecker, J, Stehle, T, Boeckler, F.M. | | Deposit date: | 2022-04-05 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Screening of a Halogen-Enriched Fragment Library Leads to Unconventional Binding Modes.
J.Med.Chem., 65, 2022
|
|
8A92
 
 | | p53-Y220C Core Domain in Complex with a Bromo-trifluoro-pyrazole-amine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-bromanyl-5-(trifluoromethyl)-1H-pyrazol-3-amine, Cellular tumor antigen p53, ... | | Authors: | Stahlecker, J, Braun, M.B, Stehle, T, Boeckler, F.M. | | Deposit date: | 2022-06-27 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Revisiting a challenging p53 binding site: a diversity-optimized HEFLib reveals diverse binding modes in T-p53C-Y220C.
Rsc Med Chem, 13, 2022
|
|
5IVN
 
 | | BC2 nanobody in complex with the BC2 peptide tag | | Descriptor: | BC2-nanobody, Cadherin derived peptide | | Authors: | Braun, M.B, Stehle, T. | | Deposit date: | 2016-03-21 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Peptides in headlock - a novel high-affinity and versatile peptide-binding nanobody for proteomics and microscopy.
Sci Rep, 6, 2016
|
|
3I4Z
 
 | | Crystal structure of the dimethylallyl tryptophan synthase FgaPT2 from Aspergillus fumigatus | | Descriptor: | 1,3-BUTANEDIOL, GLYCEROL, Tryptophan dimethylallyltransferase | | Authors: | Schall, C, Zocher, G, Stehle, T. | | Deposit date: | 2009-07-03 | | Release date: | 2009-09-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The structure of dimethylallyl tryptophan synthase reveals a common architecture of aromatic prenyltransferases in fungi and bacteria
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3I4X
 
 | | Crystal structure of the dimethylallyl tryptophan synthase FgaPT2 from Aspergillus fumigatus in complex with Trp and DMSPP | | Descriptor: | DIMETHYLALLYL S-THIOLODIPHOSPHATE, GLYCEROL, TRYPTOPHAN, ... | | Authors: | Schall, C, Zocher, G, Stehle, T. | | Deposit date: | 2009-07-03 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of dimethylallyl tryptophan synthase reveals a common architecture of aromatic prenyltransferases in fungi and bacteria
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6GS2
 
 | | Crystal Structure of the GatD/MurT Enzyme Complex from Staphylococcus aureus | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, SA1707 protein, ... | | Authors: | Muckenfuss, L.M, Noeldeke, E.R, Niemann, V, Zocher, G, Stehle, T. | | Deposit date: | 2018-06-13 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural basis of cell wall peptidoglycan amidation by the GatD/MurT complex of Staphylococcus aureus.
Sci Rep, 8, 2018
|
|
8Q6X
 
 | | Crystal structure of Cytochrome P450 GymB5 from Streptomyces katrae | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Freytag, J, Mueller, J.M, Stehle, T, Zocher, G. | | Deposit date: | 2023-08-15 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Intramolecular Coupling and Nucleobase Transfer - How Cytochrome P450 Enzymes GymBx Establish Their Chemoselectivity
Chemcatchem, 16, 2024
|
|
8Q6Y
 
 | | Crystal structure of Cytochrome P450 GymB5 from Streptomyces katrae in complex with cYY and Hypoxanthine | | Descriptor: | (3S,6S)-3,6-bis(4-hydroxybenzyl)piperazine-2,5-dione, 1,2-ETHANEDIOL, Cytochrome P450, ... | | Authors: | Freytag, J, Mueller, J.M, Stehle, T, Zocher, G. | | Deposit date: | 2023-08-15 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Intramolecular Coupling and Nucleobase Transfer - How Cytochrome P450 Enzymes GymBx Establish Their Chemoselectivity
Chemcatchem, 16, 2024
|
|
3S6Y
 
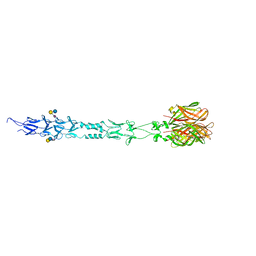 | | Structure of reovirus attachment protein sigma1 in complex with alpha-2,6-sialyllactose | | Descriptor: | N-acetyl-alpha-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Outer capsid protein sigma-1 | | Authors: | Reiter, D.M, Dermody, T.S, Stehle, T. | | Deposit date: | 2011-05-26 | | Release date: | 2011-11-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of reovirus attachment protein sigma1 in complex with sialylated oligosaccharides
Plos Pathog., 7, 2011
|
|
3S6X
 
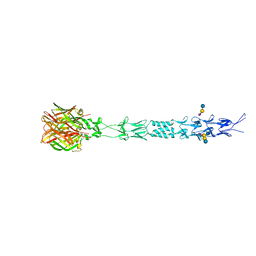 | |
3S6Z
 
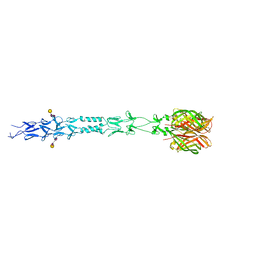 | |
7NKT
 
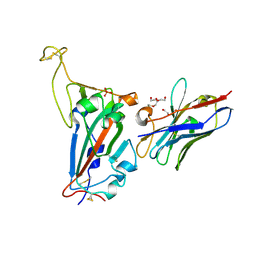 | | RBD domain of SARS-CoV2 in complex with neutralizing nanobody NM1226 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Ostertag, E, Zocher, G, Stehle, T. | | Deposit date: | 2021-02-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NeutrobodyPlex-monitoring SARS-CoV-2 neutralizing immune responses using nanobodies.
Embo Rep., 22, 2021
|
|
4MBZ
 
 | | Structure of B-Lymphotropic Polyomavirus VP1 in complex with 3'-sialyllactosamine | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Khan, Z.M, Neu, U, Stehle, T. | | Deposit date: | 2013-08-21 | | Release date: | 2013-12-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of B-Lymphotropic Polyomavirus VP1 in Complex with Oligosaccharide Ligands.
Plos Pathog., 9, 2013
|
|
7OJ7
 
 | |
4Y7T
 
 | | Structural analysis of MurU | | Descriptor: | GLYCEROL, Nucleotidyl transferase, SULFATE ION | | Authors: | Renner-Schneck, M.G, Stehle, T. | | Deposit date: | 2015-02-16 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the N-Acetylmuramic Acid alpha-1-Phosphate (MurNAc-alpha 1-P) Uridylyltransferase MurU, a Minimal Sugar Nucleotidyltransferase and Potential Drug Target Enzyme in Gram-negative Pathogens.
J.Biol.Chem., 290, 2015
|
|
4FMH
 
 | | Merkel Cell Polyomavirus VP1 in complex with Disialyllactose | | Descriptor: | CHLORIDE ION, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, ... | | Authors: | Neu, U, Hengel, H, Stehle, T. | | Deposit date: | 2012-06-17 | | Release date: | 2012-09-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of Merkel Cell Polyomavirus VP1 Complexes Define a Sialic Acid Binding Site Required for Infection.
Plos Pathog., 8, 2012
|
|
4FMI
 
 | | Merkel cell polyomavirus VP1 in complex with 3'-sialyllactosamine | | Descriptor: | CHLORIDE ION, GLYCEROL, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Neu, U, Hengel, H, Stehle, T. | | Deposit date: | 2012-06-17 | | Release date: | 2012-09-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Merkel Cell Polyomavirus VP1 Complexes Define a Sialic Acid Binding Site Required for Infection.
Plos Pathog., 8, 2012
|
|
3MOR
 
 | | Crystal structure of Cathepsin B from Trypanosoma Brucei | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Cathepsin B-like cysteine protease, ... | | Authors: | Cupelli, K, Stehle, T. | | Deposit date: | 2010-04-23 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | In vivo protein crystallization opens new routes in structural biology.
Nat.Methods, 9, 2012
|
|
4Y7U
 
 | | Structural analysis of MurU | | Descriptor: | 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, GLYCEROL, ... | | Authors: | Renner-Schneck, M.G, Stehle, T. | | Deposit date: | 2015-02-16 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the N-Acetylmuramic Acid alpha-1-Phosphate (MurNAc-alpha 1-P) Uridylyltransferase MurU, a Minimal Sugar Nucleotidyltransferase and Potential Drug Target Enzyme in Gram-negative Pathogens.
J.Biol.Chem., 290, 2015
|
|
4Y7V
 
 | | Structural analysis of MurU | | Descriptor: | 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, GLYCEROL, IMIDODIPHOSPHORIC ACID, ... | | Authors: | Renner-Schneck, M.G, Stehle, T. | | Deposit date: | 2015-02-16 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the N-Acetylmuramic Acid alpha-1-Phosphate (MurNAc-alpha 1-P) Uridylyltransferase MurU, a Minimal Sugar Nucleotidyltransferase and Potential Drug Target Enzyme in Gram-negative Pathogens.
J.Biol.Chem., 290, 2015
|
|
4FMG
 
 | |
4FMJ
 
 | | Merkel cell polyomavirus VP1 in complex with GD1a oligosaccharide | | Descriptor: | CHLORIDE ION, GLYCEROL, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Neu, U, Hengel, H, Stehle, T. | | Deposit date: | 2012-06-17 | | Release date: | 2012-09-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Merkel Cell Polyomavirus VP1 Complexes Define a Sialic Acid Binding Site Required for Infection.
Plos Pathog., 8, 2012
|
|
4MBY
 
 | | Structure of B-Lymphotropic Polyomavirus VP1 in complex with 3'-sialyllactose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Khan, Z.M, Neu, U, Stehle, T. | | Deposit date: | 2013-08-21 | | Release date: | 2013-12-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structures of B-Lymphotropic Polyomavirus VP1 in Complex with Oligosaccharide Ligands.
Plos Pathog., 9, 2013
|
|
4L68
 
 | |
