1WDU
 
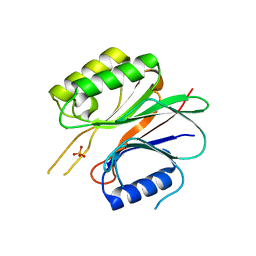 | | Endonuclease domain of TRAS1, a telomere-specific non-LTR retrotransposon | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, TRAS1 ORF2p | | Authors: | Maita, N, Anzai, T, Aoyagi, H, Mizuno, H, Fujiwara, H. | | Deposit date: | 2004-05-17 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the endonuclease domain encoded by the telomere-specific long interspersed nuclear element, TRAS1
J.Biol.Chem., 279, 2004
|
|
5AXE
 
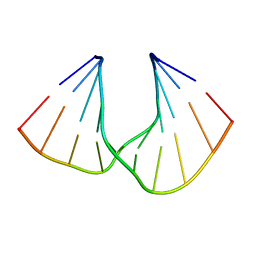 | | Crystal Structure Analysis of DNA Duplexes containing sulfoamide-bridged nucleic acid (SuNA-NH) | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(LSH)P*AP*CP*GP*C)-3') | | Authors: | Mitsuoka, Y, Aoyama, H, Kugimiya, A, Fujimura, Y, Yamamoto, T, Waki, R, Wada, F, Tahara, S, Sawamura, M, Noda, M, Hari, Y, Obika, S. | | Deposit date: | 2015-07-28 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Effect of an N-substituent in sulfonamide-bridged nucleic acid (SuNA) on hybridization ability and duplex structure.
Org.Biomol.Chem., 14, 2016
|
|
5AXF
 
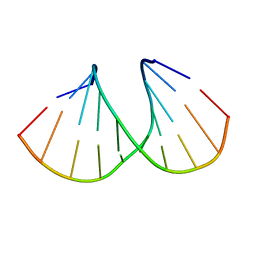 | | Crystal Structure Analysis of DNA Duplexes containing sulfoamide-bridged nucleic acid (SuNA-NMe) | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(LSM)P*AP*CP*GP*C)-3') | | Authors: | Mitsuoka, Y, Aoyama, H, Kugimiya, A, Fujimura, Y, Yamamoto, T, Waki, R, Wada, F, Tahara, S, Sawamura, M, Noda, M, Hari, Y, Obika, S. | | Deposit date: | 2015-07-28 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Effect of an N-substituent in sulfonamide-bridged nucleic acid (SuNA) on hybridization ability and duplex structure.
Org.Biomol.Chem., 14, 2016
|
|
1KV9
 
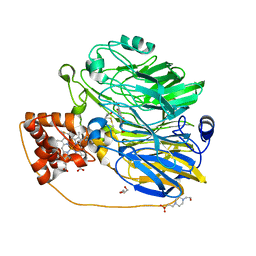 | | Structure at 1.9 A Resolution of a Quinohemoprotein Alcohol Dehydrogenase from Pseudomonas putida HK5 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETONE, CALCIUM ION, ... | | Authors: | Chen, Z.-W, Matsushita, K, Yamashita, T, Fujii, T, Toyama, H, Adachi, O, Bellamy, H.D, Mathews, F.S. | | Deposit date: | 2002-01-25 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure at 1.9 A resolution of a quinohemoprotein alcohol dehydrogenase from Pseudomonas putida HK5.
Structure, 10, 2002
|
|
8H8L
 
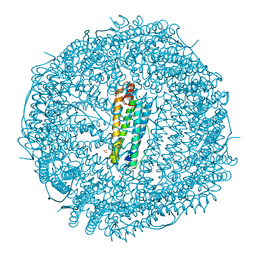 | | Crystal structure of apo-R52F/E56F/R59F/E63F-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Hishikawa, Y, Noya, H, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2022-10-23 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidating Conformational Dynamics and Thermostability of Designed Aromatic Clusters by Using Protein Cages.
Chemistry, 29, 2023
|
|
8H8N
 
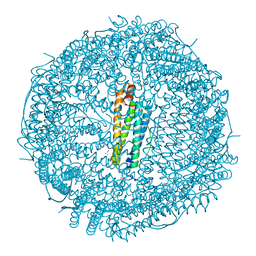 | | Crystal structure of apo-R52Y/E56Y/R59Y/E63Y-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Hishikawa, Y, Noya, H, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2022-10-23 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidating Conformational Dynamics and Thermostability of Designed Aromatic Clusters by Using Protein Cages.
Chemistry, 29, 2023
|
|
8H8M
 
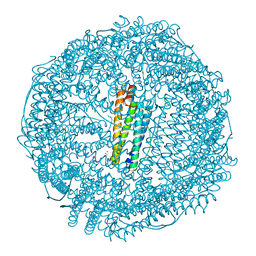 | | Crystal structure of apo-E53F/E57F/E60F/E64F-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Hishikawa, Y, Noya, H, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2022-10-23 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidating Conformational Dynamics and Thermostability of Designed Aromatic Clusters by Using Protein Cages.
Chemistry, 29, 2023
|
|
8H8O
 
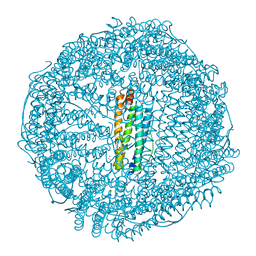 | | Crystal structure of apo-R52W/E56W/R59W/E63W-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Hishikawa, Y, Noya, H, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2022-10-23 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidating Conformational Dynamics and Thermostability of Designed Aromatic Clusters by Using Protein Cages.
Chemistry, 29, 2023
|
|
1V55
 
 | | Bovine heart cytochrome c oxidase at the fully reduced state | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Tsukihara, T, Shimokata, K, Katayama, Y, Shimada, H, Muramoto, K, Aoyama, H, Mochizuki, M, Shinzawa-Itoh, K, Yamashita, E, Yao, M, Ishimura, Y, Yoshikawa, S. | | Deposit date: | 2003-11-21 | | Release date: | 2003-12-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The low-spin heme of cytochrome c oxidase as the driving element of the proton-pumping process.
Proc.Natl.Acad.Sci.Usa, 100, 2003
|
|
1V54
 
 | | Bovine heart cytochrome c oxidase at the fully oxidized state | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Tsukihara, T, Shimokata, K, Katayama, Y, Shimada, H, Muramoto, K, Aoyama, H, Mochizuki, M, Shinzawa-Itoh, K, Yamashita, E, Yao, M, Ishimura, Y, Yoshikawa, S. | | Deposit date: | 2003-11-21 | | Release date: | 2003-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The low-spin heme of cytochrome c oxidase as the driving element of the proton-pumping process.
Proc.Natl.Acad.Sci.Usa, 100, 2003
|
|
1SIO
 
 | | Structure of Kumamolisin-As complexed with a covalently-bound inhibitor, AcIPF | | Descriptor: | Ace-ILE-PRO-PHL peptide inhibitor, CALCIUM ION, SULFATE ION, ... | | Authors: | Li, M, Wlodawer, A, Gustchina, A, Tsuruoka, N, Ashida, M, Minakata, H, Oyama, H, Oda, K, Nishino, T, Nakayama, T. | | Deposit date: | 2004-03-01 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic and biochemical investigations of kumamolisin-As, a serine-carboxyl peptidase with collagenase activity
J.Biol.Chem., 279, 2004
|
|
1SIU
 
 | | KUMAMOLISIN-AS E78H MUTANT | | Descriptor: | CALCIUM ION, SULFATE ION, kumamolisin-As | | Authors: | Li, M, Wlodawer, A, Gustchina, A, Tsuruoka, N, Ashida, M, Minakata, H, Oyama, H, Oda, K, Nishino, T, Nakayama, T. | | Deposit date: | 2004-03-01 | | Release date: | 2004-03-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystallographic and biochemical investigations of kumamolisin-As, a serine-carboxyl peptidase with collagenase activity
J.Biol.Chem., 279, 2004
|
|
7DRU
 
 | |
7WT1
 
 | |
7WT0
 
 | | human glyoxalase I (with C-ter His tag) in complex with TLSC702 | | Descriptor: | (~{E})-3-(1,3-benzothiazol-2-yl)-4-(4-methoxyphenyl)but-3-enoic acid, Lactoylglutathione lyase, ZINC ION | | Authors: | Usami, M, Yokoyama, H. | | Deposit date: | 2022-02-03 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of human glyoxalase I and its complex with TLSC702 reveal inhibitor binding mode and substrate preference.
Febs Lett., 596, 2022
|
|
7WSZ
 
 | | human glyoxalase I (with C-ter His tag) in glycerol-bound form | | Descriptor: | BETA-MERCAPTOETHANOL, BORIC ACID, GLYCEROL, ... | | Authors: | Usami, M, Ando, K, Yokoyama, H. | | Deposit date: | 2022-02-03 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structures of human glyoxalase I and its complex with TLSC702 reveal inhibitor binding mode and substrate preference.
Febs Lett., 596, 2022
|
|
7WT2
 
 | | human glyoxalase I in complex with TLSC702 | | Descriptor: | (~{E})-3-(1,3-benzothiazol-2-yl)-4-(4-methoxyphenyl)but-3-enoic acid, Lactoylglutathione lyase, ZINC ION | | Authors: | Usami, M, Yokoyama, H. | | Deposit date: | 2022-02-03 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of human glyoxalase I and its complex with TLSC702 reveal inhibitor binding mode and substrate preference.
Febs Lett., 596, 2022
|
|
2EI9
 
 | | Crystal structure of R1Bm endonuclease domain | | Descriptor: | ACETIC ACID, Non-LTR retrotransposon R1Bmks ORF2 protein | | Authors: | Maita, N, Aoyagi, H, Osanai, M, Shirakawa, M, Fujiwara, H. | | Deposit date: | 2007-03-12 | | Release date: | 2007-07-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the sequence specificity of the R1Bm endonuclease domain by structural and biochemical studies.
Nucleic Acids Res., 35, 2007
|
|
2DYS
 
 | | Bovine heart cytochrome C oxidase modified by DCCD | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shinzawa-Itoh, K, Aoyama, H, Muramoto, K, Kurauchi, T, Mizushima, T, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2006-09-16 | | Release date: | 2007-04-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase
Embo J., 26, 2007
|
|
2DYR
 
 | | Bovine heart cytochrome C oxidase at the fully oxidized state | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shinzawa-Itoh, K, Aoyama, H, Muramoto, K, Kurauchi, T, Mizushima, T, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2006-09-16 | | Release date: | 2007-04-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase
Embo J., 26, 2007
|
|
1T1I
 
 | | High Resolution Crystal Structure of Mutant W129A of Kumamolisin, a Sedolisin Type Proteinase (previously called Kumamolysin or KSCP) | | Descriptor: | CALCIUM ION, SULFATE ION, kumamolisin | | Authors: | Comellas-Bigler, M, Maskos, K, Huber, R, Oyama, H, Oda, K, Bode, W. | | Deposit date: | 2004-04-16 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | 1.2 a crystal structure of the serine carboxyl proteinase pro-kumamolisin: structure of an intact pro-subtilase
Structure, 12, 2004
|
|
1S2K
 
 | | Structure of SCP-B a member of the Eqolisin family of Peptidases in a complex with a Tripeptide Ala-Ile-His | | Descriptor: | Ala-Ile-His tripeptide, Scytalidopepsin B, TYROSINE | | Authors: | Fujinaga, M, Cherney, M.M, Oyama, H, Oda, K, James, M.N. | | Deposit date: | 2004-01-08 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The molecular structure and catalytic mechanism of a novel carboxyl peptidase from Scytalidium lignicolum
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1T1G
 
 | | High Resolution Crystal Structure of Mutant E23A of Kumamolisin, a sedolisin type proteinase (previously called Kumamolysin or KSCP) | | Descriptor: | CALCIUM ION, SULFATE ION, kumamolisin | | Authors: | Comellas-Bigler, M, Maskos, K, Huber, R, Oyama, H, Oda, K, Bode, W. | | Deposit date: | 2004-04-16 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | 1.2 a crystal structure of the serine carboxyl proteinase pro-kumamolisin: structure of an intact pro-subtilase
Structure, 12, 2004
|
|
1S2B
 
 | | Structure of SCP-B the first member of the Eqolisin family of Peptidases to have its structure determined | | Descriptor: | Scytalidopepsin B | | Authors: | Fujinaga, M, Cherney, M.M, Oyama, H, Oda, K, James, M.N. | | Deposit date: | 2004-01-08 | | Release date: | 2004-04-27 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The molecular structure and catalytic mechanism of a novel carboxyl peptidase from Scytalidium lignicolum
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1T1E
 
 | | High Resolution Crystal Structure of the Intact Pro-Kumamolisin, a Sedolisin Type Proteinase (previously called Kumamolysin or KSCP) | | Descriptor: | CALCIUM ION, kumamolisin | | Authors: | Comellas-Bigler, M, Maskos, K, Huber, R, Oyama, H, Oda, K, Bode, W. | | Deposit date: | 2004-04-16 | | Release date: | 2004-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | 1.2 a crystal structure of the serine carboxyl proteinase pro-kumamolisin: structure of an intact pro-subtilase
Structure, 12, 2004
|
|
