8EPG
 
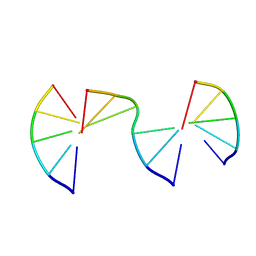 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*C)-3'), DNA (5'-D(P*GP*CP*CP*GP*C)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-10-05 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
8F42
 
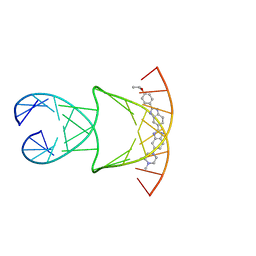 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | 2'-(4-ETHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*TP*TP*AP*AP*GP*GP*AP*AP*TP*TP*CP*GP*C)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
8EP8
 
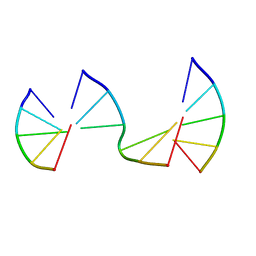 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*GP*C)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-10-05 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
8F40
 
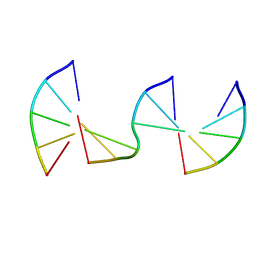 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*T)-3'), DNA (5'-D(P*AP*AP*GP*GP*AP*A)-3'), DNA (5'-D(P*TP*TP*CP*GP*C)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
8EPF
 
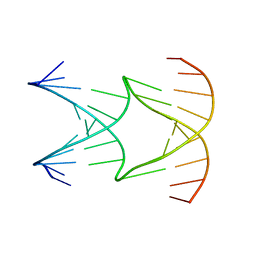 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | DNA (5'-D(*AP*CP*GP*CP*TP*GP*GP*TP*GP*GP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*GP*TP*AP*CP*CP*AP*GP*CP*CP*GP*AP*AP*CP*CP*TP*G)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-10-05 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
4KW8
 
 | |
4KW9
 
 | |
4KW4
 
 | |
3SLO
 
 | | Pre-cleavage Structure of the Autotransporter EspP - N1023D mutant | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Serine protease espP | | Authors: | Barnard, T.B, Noinaj, N, Easley, N.C, Kuszak, A.J, Buchanan, S.K. | | Deposit date: | 2011-06-24 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Molecular basis for the activation of a catalytic asparagine residue in a self-cleaving bacterial autotransporter.
J.Mol.Biol., 415, 2012
|
|
3SLT
 
 | | Pre-cleavage Structure of the Autotransporter EspP - N1023S Mutant | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Serine protease espP | | Authors: | Barnard, T.B, Noinaj, N, Easley, N.C, Kuszak, A.J, Buchanan, S.K. | | Deposit date: | 2011-06-25 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Molecular basis for the activation of a catalytic asparagine residue in a self-cleaving bacterial autotransporter.
J.Mol.Biol., 415, 2012
|
|
3QRC
 
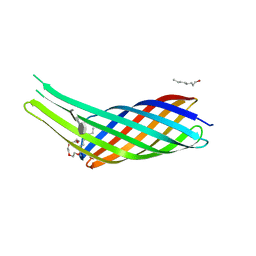 | | The crystal structure of Ail, the attachment invasion locus protein of Yersinia pestis, in complex with the heparin analogue sucrose octasulfate | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, Attachment invasion locus protein | | Authors: | Yamashita, S, Lukacik, P, Noinaj, N, Buchanan, S.K. | | Deposit date: | 2011-02-17 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Structural Insights into Ail-Mediated Adhesion in Yersinia pestis.
Structure, 19, 2011
|
|
3QRA
 
 | | The crystal structure of Ail, the attachment invasion locus protein of Yersinia pestis | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Attachment invasion locus protein | | Authors: | Yamashita, S, Lukacik, P, Noinaj, N, Buchanan, S.K. | | Deposit date: | 2011-02-17 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural Insights into Ail-Mediated Adhesion in Yersinia pestis.
Structure, 19, 2011
|
|
7UW6
 
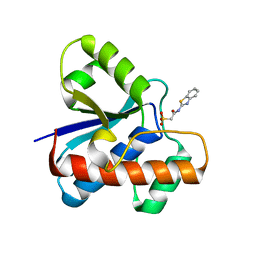 | | The co-crystal structure of low molecular weight protein tyrosine phosphatase (LMW-PTP) with a small molecule inhibitor SPAA-2 | | Descriptor: | 2-[(1,3-benzothiazol-2-yl)amino]-2-oxoethane-1-sulfonic acid, Low molecular weight phosphotyrosine protein phosphatase | | Authors: | Wang, J, Zhang, Z.Y. | | Deposit date: | 2022-05-02 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Design of Active-Site-Directed, Highly Potent, Selective, and Orally Bioavailable Low-Molecular-Weight Protein Tyrosine Phosphatase Inhibitors.
J.Med.Chem., 65, 2022
|
|
4EPA
 
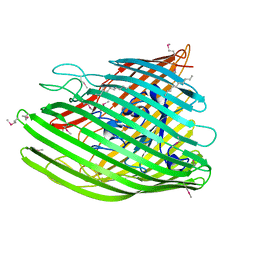 | |
4EPF
 
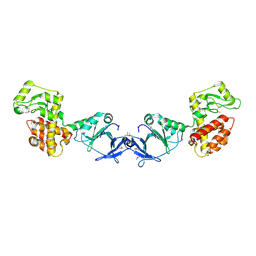 | |
7TAW
 
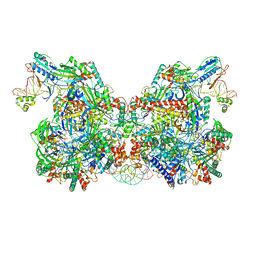 | | Cryo-EM structure of the Csy-AcrIF24-promoter DNA dimer | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR type I-F/YPEST-associated protein Csy3, ... | | Authors: | Mukherjee, I.A, Chang, L. | | Deposit date: | 2021-12-21 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of AcrIF24 as an anti-CRISPR protein and transcriptional suppressor.
Nat.Chem.Biol., 18, 2022
|
|
7TAX
 
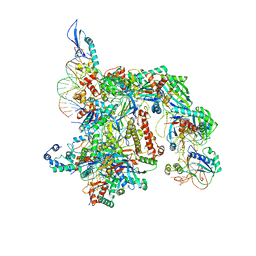 | | Cryo-EM structure of the Csy-AcrIF24-promoter DNA complex | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR type I-F/YPEST-associated protein Csy3, ... | | Authors: | Mukherjee, I.A, Chang, L. | | Deposit date: | 2021-12-21 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of AcrIF24 as an anti-CRISPR protein and transcriptional suppressor.
Nat.Chem.Biol., 18, 2022
|
|
7T3K
 
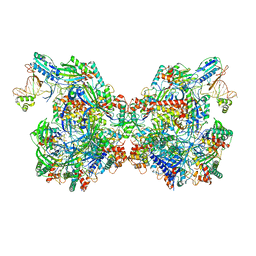 | | Cryo-EM structure of Csy-AcrIF24 dimer | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR type I-F/YPEST-associated protein Csy3, ... | | Authors: | Mukherjee, I.A, Chang, L. | | Deposit date: | 2021-12-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of AcrIF24 as an anti-CRISPR protein and transcriptional suppressor.
Nat.Chem.Biol., 18, 2022
|
|
7T3J
 
 | | Cryo-EM structure of Csy-AcrIF24 | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR type I-F/YPEST-associated protein Csy3, ... | | Authors: | Mukherjee, I.A, Chang, L. | | Deposit date: | 2021-12-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of AcrIF24 as an anti-CRISPR protein and transcriptional suppressor.
Nat.Chem.Biol., 18, 2022
|
|
7T3L
 
 | | Cryo-EM structure of Csy-AcrIF24-DNA dimer | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR type I-F/YPEST-associated protein Csy3, ... | | Authors: | Mukherjee, I.A, Chang, L. | | Deposit date: | 2021-12-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of AcrIF24 as an anti-CRISPR protein and transcriptional suppressor.
Nat.Chem.Biol., 18, 2022
|
|
