1IE9
 
 | | Crystal Structure Of The Nuclear Receptor For Vitamin D Ligand Binding Domain Bound to MC1288 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, VITAMIN D3 RECEPTOR | | Authors: | Tocchini-Valentini, G, Rochel, N, Wurtz, J.M, Mitschler, A, Moras, D. | | Deposit date: | 2001-04-09 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of the vitamin D receptor complexed to superagonist 20-epi ligands.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
4XZH
 
 | | Crystal structure of human Aldose Reductase complexed with NADP+ and JF0048 | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [3-(4-chloro-3-nitrobenzyl)-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl]acetic acid | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Dominguez, M, de Lera, A.R, Farres, J, Pares, X, Podjarny, A. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
4XZI
 
 | | Crystal structure of human Aldose Reductase complexed with NADP+ and JF0049 | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [2,4-dioxo-3-(2,3,4,5-tetrabromo-6-methoxybenzyl)-3,4-dihydropyrimidin-1(2H)-yl]acetic acid | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Dominguez, M, de Lera, A.R, Farres, J, Pares, X, Podjarny, A. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
4XZN
 
 | | Crystal structure of the methylated K125R/V301L AKR1B10 Holoenzyme | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Podjarny, A. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
1H3O
 
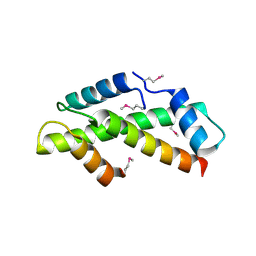 | | Crystal Structure of the Human TAF4-TAF12 (TAFII135-TAFII20) Complex | | Descriptor: | TRANSCRIPTION INITIATION FACTOR TFIID 135 KDA SUBUNIT, TRANSCRIPTION INITIATION FACTOR TFIID 20/15 KDA SUBUNITS | | Authors: | Werten, S, Mitschler, A, Moras, D. | | Deposit date: | 2002-09-12 | | Release date: | 2002-09-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of a Subcomplex of Human Transcription Factor TFIID Formed by TATA Binding Protein-Associated Factors Htaf4 (Htaf(II)135) and Htaf12 (Htaf(II)20).
J.Biol.Chem., 277, 2002
|
|
4XZL
 
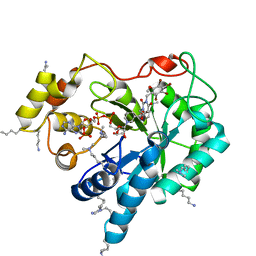 | | Crystal structure of human AKR1B10 complexed with NADP+ and JF0049 | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Dominguez, M, de Lera, A.R, Farres, J, Pares, X, Podjarny, A. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
4XZM
 
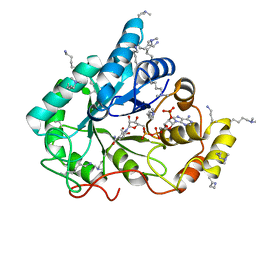 | | Crystal structure of the methylated wild-type AKR1B10 holoenzyme | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Podjarny, A. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
4WKA
 
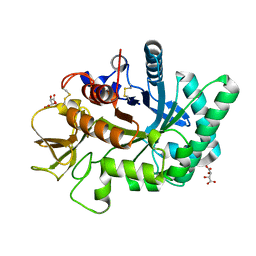 | | Crystal structure of human chitotriosidase-1 catalytic domain at 0.95 A resolution | | Descriptor: | Chitotriosidase-1, L(+)-TARTARIC ACID | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5OUJ
 
 | | Crystal structure of human AKR1B1 complexed with NADP+ and compound 39 | | Descriptor: | 2-[(1~{R})-5-(4-chlorophenyl)-9-fluoranyl-3-methyl-1-oxidanyl-1~{H}-pyrimido[4,5-c]quinolin-2-yl]ethanoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Metwally, K, Podjarny, A. | | Deposit date: | 2017-08-24 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Design, synthesis, structure-activity relationships and X-ray structural studies of novel 1-oxopyrimido[4,5-c]quinoline-2-acetic acid derivatives as selective and potent inhibitors of human aldose reductase.
Eur J Med Chem, 152, 2018
|
|
5OUK
 
 | | Crystal structure of human AKR1B1 complexed with NADP+ and compound 41 | | Descriptor: | 2-[9-fluoranyl-5-(4-methoxyphenyl)-3-methyl-1-oxidanylidene-pyrimido[4,5-c]quinolin-2-yl]ethanoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Metwally, K, Podjarny, A. | | Deposit date: | 2017-08-24 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.959 Å) | | Cite: | Design, synthesis, structure-activity relationships and X-ray structural studies of novel 1-oxopyrimido[4,5-c]quinoline-2-acetic acid derivatives as selective and potent inhibitors of human aldose reductase.
Eur J Med Chem, 152, 2018
|
|
5OU0
 
 | | Crystal structure of human AKR1B1 complexed with NADP+ and compound 37 | | Descriptor: | 2-[5-(4-chlorophenyl)-3-methyl-1-oxidanylidene-pyrimido[4,5-c]quinolin-2-yl]ethanoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Metwally, K, Podjarny, A. | | Deposit date: | 2017-08-23 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Design, synthesis, structure-activity relationships and X-ray structural studies of novel 1-oxopyrimido[4,5-c]quinoline-2-acetic acid derivatives as selective and potent inhibitors of human aldose reductase.
Eur J Med Chem, 152, 2018
|
|
4WKH
 
 | | Crystal structure of human chitotriosidase-1 catalytic domain in complex with chitobiose (1mM) at 1.05 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitotriosidase-1 | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WEV
 
 | | Crystal structure of human AKR1B10 complexed with NADP+ and sulindac | | Descriptor: | Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [(1Z)-5-fluoro-2-methyl-1-{4-[methylsulfinyl]benzylidene}-1H-inden-3-yl]acetic acid | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Crespo, I, Porte, S, Pares, X, Farres, J, Podjarny, A. | | Deposit date: | 2014-09-11 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.453 Å) | | Cite: | Structural analysis of sulindac as an inhibitor of aldose reductase and AKR1B10.
Chem.Biol.Interact., 234, 2015
|
|
4WKF
 
 | | Crystal structure of human chitotriosidase-1 catalytic domain in complex with chitobiose (2.5mM) at 1.10 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitotriosidase-1 | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1IE8
 
 | | Crystal Structure Of The Nuclear Receptor For Vitamin D Ligand Binding Domain Bound to KH1060 | | Descriptor: | 5-(2-{1-[1-(4-ETHYL-4-HYDROXY-HEXYLOXY)-ETHYL]-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE}-ETHYLIDENE)-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, VITAMIN D3 RECEPTOR | | Authors: | Tocchini-Valentini, G, Rochel, N, Wurtz, J.M, Mitschler, A, Moras, D. | | Deposit date: | 2001-04-09 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structures of the vitamin D receptor complexed to superagonist 20-epi ligands.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1R20
 
 | | Crystal structure of the ligand-binding domains of the heterodimer EcR/USP bound to the synthetic agonist BYI06830 | | Descriptor: | ECDYSONE RECEPTOR, L-ALPHA-PHOSPHATIDYL-BETA-OLEOYL-GAMMA-PALMITOYL-PHOSPHATIDYLETHANOLAMINE, N-(TERT-BUTYL)-3,5-DIMETHYL-N'-[(5-METHYL-2,3-DIHYDRO-1,4-BENZODIOXIN-6-YL)CARBONYL]BENZOHYDRAZIDE, ... | | Authors: | Billas, I.M.L, Iwema, T, Garnier, J.M, Mitschler, A, Rochel, N, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-09-25 | | Release date: | 2003-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural adaptability in the ligand-binding pocket of the ecdysone hormone receptor.
Nature, 426, 2003
|
|
1R1K
 
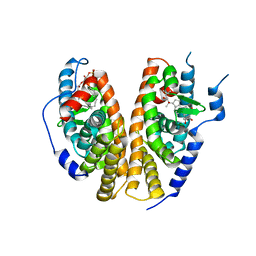 | | Crystal structure of the ligand-binding domains of the heterodimer EcR/USP bound to ponasterone A | | Descriptor: | 2,3,14,20,22-PENTAHYDROXYCHOLEST-7-EN-6-ONE, Ecdysone receptor, L-ALPHA-PHOSPHATIDYL-BETA-OLEOYL-GAMMA-PALMITOYL-PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Billas, I.M.L, Iwema, T, Garnier, J.-M, Mitschler, A, Rochel, N, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-09-24 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural adaptability in the ligand-binding pocket of the ecdysone hormone receptor.
Nature, 426, 2003
|
|
1BY3
 
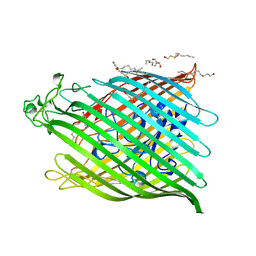 | | FHUA FROM E. COLI | | Descriptor: | N-OCTYL-2-HYDROXYETHYL SULFOXIDE, PROTEIN (FERRICHROME-IRON RECEPTOR PRECURSOR (FHUA)) | | Authors: | Locher, K.P, Rees, B, Koebnik, R, Mitschler, A, Moulinier, L, Rosenbusch, J.P, Moras, D. | | Deposit date: | 1998-10-22 | | Release date: | 1999-01-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes.
Cell(Cambridge,Mass.), 95, 1998
|
|
1BY5
 
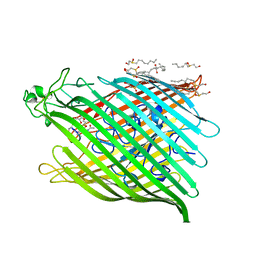 | | FHUA FROM E. COLI, WITH ITS LIGAND FERRICHROME | | Descriptor: | FE (III) ION, FERRIC HYDROXAMATE UPTAKE PROTEIN, FERRICHROME, ... | | Authors: | Locher, K.P, Rees, B, Koebnik, R, Mitschler, A, Moulinier, L, Rosenbusch, J.P, Moras, D. | | Deposit date: | 1998-10-23 | | Release date: | 1999-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes.
Cell(Cambridge,Mass.), 95, 1998
|
|
3MTY
 
 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region A of the crystal. First step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-05-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3MU1
 
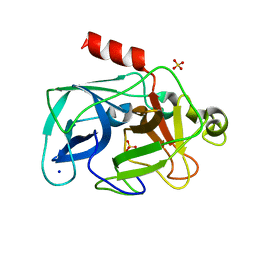 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region A of the crystal. Fifth step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3MU0
 
 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region A of the crystal. Third step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3MU8
 
 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region B of the crystal. Fifth step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-05-02 | | Release date: | 2010-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3MU4
 
 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region B of the crystal. First step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-05-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3MU5
 
 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region B of the crystal. Third step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
