1STM
 
 | | SATELLITE PANICUM MOSAIC VIRUS | | Descriptor: | SATELLITE PANICUM MOSAIC VIRUS | | Authors: | Ban, N, McPherson, A. | | Deposit date: | 1995-07-12 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of satellite panicum mosaic virus at 1.9 A resolution.
Nat.Struct.Biol., 2, 1995
|
|
1AUY
 
 | |
5CVZ
 
 | |
5CW0
 
 | |
1DDL
 
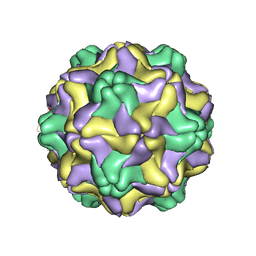 | | DESMODIUM YELLOW MOTTLE TYMOVIRUS | | Descriptor: | DESMODIUM YELLOW MOTTLE VIRUS, RNA (5'-R(P*UP*U)-3'), RNA (5'-R(P*UP*UP*UP*UP*UP*UP*U)-3') | | Authors: | Larson, S.B, Day, J, Canady, M.A, Greenwood, A, McPherson, A. | | Deposit date: | 1999-11-10 | | Release date: | 2000-10-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Refined structure of desmodium yellow mottle tymovirus at 2.7 A resolution.
J.Mol.Biol., 301, 2000
|
|
1NOF
 
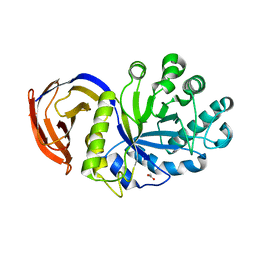 | | THE FIRST CRYSTALLOGRAPHIC STRUCTURE OF A XYLANASE FROM GLYCOSYL HYDROLASE FAMILY 5: IMPLICATIONS FOR CATALYSIS | | Descriptor: | ACETATE ION, xylanase | | Authors: | Larson, S.B, Day, J, McPherson, A, Barba De La Rosa, A.P, Keen, N.T. | | Deposit date: | 2003-01-16 | | Release date: | 2003-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | First crystallographic structure of a xylanase from glycoside hydrolase family 5: implications for catalysis.
Biochemistry, 42, 2003
|
|
1RTB
 
 | |
6C75
 
 | |
5TW0
 
 | |
6C7L
 
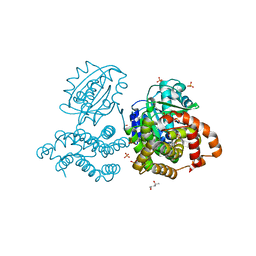 | | Structure of Iron containing alcohol dehydrogenase from Thermococcus thioreducens in a tetragonal crystal form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Jones, J.A, Larson, S.B, McPherson, A. | | Deposit date: | 2018-01-22 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The structure of an iron-containing alcohol dehydrogenase from a hyperthermophilic archaeon in two chemical states.
Acta Crystallogr F Struct Biol Commun, 75, 2019
|
|
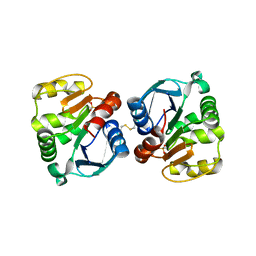
5TXW
 
 | |
6C76
 
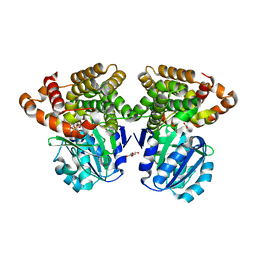 | |
1RTA
 
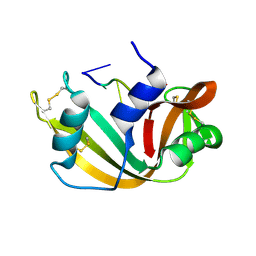 | |
1THU
 
 | | THE STRUCTURES OF THREE CRYSTAL FORMS OF THE SWEET PROTEIN THAUMATIN | | Descriptor: | THAUMATIN ISOFORM B | | Authors: | Ko, T.-P, Day, J, Greenwood, A, McPherson, A. | | Deposit date: | 1994-06-10 | | Release date: | 1994-12-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of three crystal forms of the sweet protein thaumatin.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1THV
 
 | | THE STRUCTURES OF THREE CRYSTAL FORMS OF THE SWEET PROTEIN THAUMATIN | | Descriptor: | THAUMATIN ISOFORM A | | Authors: | Ko, T.-P, Day, J, Greenwood, A, McPherson, A. | | Deposit date: | 1994-06-10 | | Release date: | 1994-12-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of three crystal forms of the sweet protein thaumatin.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1THW
 
 | | THE STRUCTURES OF THREE CRYSTAL FORMS OF THE SWEET PROTEIN THAUMATIN | | Descriptor: | L(+)-TARTARIC ACID, THAUMATIN | | Authors: | Ko, T.-P, Day, J, Greenwood, A, McPherson, A. | | Deposit date: | 1994-06-10 | | Release date: | 1994-12-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of three crystal forms of the sweet protein thaumatin.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1GHF
 
 | | ANTI-ANTI-IDIOTYPE GH1002 FAB FRAGMENT | | Descriptor: | ANTI-ANTI-IDIOTYPE GH1002 FAB FRAGMENT | | Authors: | Ban, N, Day, J, Wang, X, Ferrone, S, McPherson, A. | | Deposit date: | 1995-11-30 | | Release date: | 1996-12-23 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of an anti-anti-idiotype shows it to be self-complementary.
J.Mol.Biol., 255, 1996
|
|
1RBJ
 
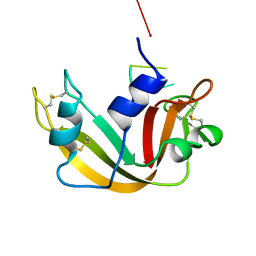 | |
3LIP
 
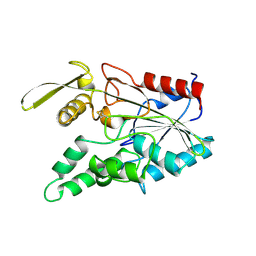 | |
2CAU
 
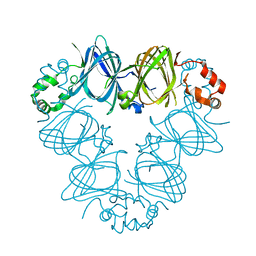 | | CANAVALIN FROM JACK BEAN | | Descriptor: | PROTEIN (CANAVALIN) | | Authors: | Ko, T.-P, Day, J, Macpherson, A. | | Deposit date: | 1998-11-20 | | Release date: | 1998-11-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The refined structure of canavalin from jack bean in two crystal forms at 2.1 and 2.0 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
2CAV
 
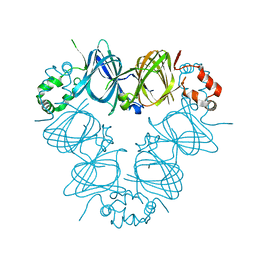 | | CANAVALIN FROM JACK BEAN | | Descriptor: | PROTEIN (CANAVALIN) | | Authors: | Ko, T.-P, Day, J, Macpherson, A. | | Deposit date: | 1998-11-20 | | Release date: | 1998-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The refined structure of canavalin from jack bean in two crystal forms at 2.1 and 2.0 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
2LIP
 
 | |
5LIP
 
 | | PSEUDOMONAS LIPASE COMPLEXED WITH RC-(RP, SP)-1,2-DIOCTYLCARBAMOYLGLYCERO-3-O-OCTYLPHOSPHONATE | | Descriptor: | CALCIUM ION, OCTYL-PHOSPHINIC ACID 1,2-BIS-OCTYLCARBAMOYLOXY-ETHYL ESTER, TRIACYL-GLYCEROL HYDROLASE | | Authors: | Lang, D.A, Dijkstra, B.W. | | Deposit date: | 1997-09-02 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of the chiral selectivity of Pseudomonas cepacia lipase
Eur.J.Biochem., 254, 1998
|
|
1SCH
 
 | | PEANUT PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PEANUT PEROXIDASE, ... | | Authors: | Schuller, D.J, Poulos, T.L. | | Deposit date: | 1996-01-23 | | Release date: | 1996-07-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The crystal structure of peanut peroxidase.
Structure, 4, 1996
|
|
1EFM
 
 | |
