6SR5
 
 | | X-ray pump X-ray probe on lysozyme.Gd nanocrystals: 102 fs time delay | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Kloos, M, Gorel, A, Nass, K. | | Deposit date: | 2019-09-04 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural dynamics in proteins induced by and probed with X-ray free-electron laser pulses.
Nat Commun, 11, 2020
|
|
6SRP
 
 | |
6SR3
 
 | | X-ray pump X-ray probe on lysozyme.Gd nanocrystals: 62 fs time delay | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Kloos, M, Gorel, A, Nass, K. | | Deposit date: | 2019-09-04 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural dynamics in proteins induced by and probed with X-ray free-electron laser pulses.
Nat Commun, 11, 2020
|
|
8U0V
 
 | | S. cerevisiae Pex1/Pex6 with 1 mM ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Peroxisomal ATPase PEX1, Peroxisomal ATPase PEX6 | | Authors: | Gardner, B.M. | | Deposit date: | 2023-08-29 | | Release date: | 2023-12-13 | | Last modified: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | The N1 domain of the peroxisomal AAA-ATPase Pex6 is required for Pex15 binding and proper assembly with Pex1.
J.Biol.Chem., 300, 2023
|
|
6DUQ
 
 | | Structure of a Rho-NusG KOW domain complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Berger, J.M, Lawson, M.R. | | Deposit date: | 2018-06-21 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Mechanism for the Regulated Control of Bacterial Transcription Termination by a Universal Adaptor Protein.
Mol. Cell, 71, 2018
|
|
5DU0
 
 | |
8U0X
 
 | | Yeast Pex6 N1(1-184) Domain | | Descriptor: | Peroxisomal ATPase PEX6 | | Authors: | Gardner, B.M, Ali, B.A. | | Deposit date: | 2023-08-29 | | Release date: | 2023-11-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The N1 domain of the peroxisomal AAA-ATPase Pex6 is required for Pex15 binding and proper assembly with Pex1.
J.Biol.Chem., 300, 2023
|
|
5DTZ
 
 | |
5VXV
 
 | | Peroxisomal membrane protein PEX15 | | Descriptor: | Peroxisomal membrane protein PEX15 | | Authors: | Gardner, B.M, Castanzo, D.T. | | Deposit date: | 2017-05-24 | | Release date: | 2018-01-17 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The peroxisomal AAA-ATPase Pex1/Pex6 unfolds substrates by processive threading.
Nat Commun, 9, 2018
|
|
2MRW
 
 | | Solution Structure of MciZ from Bacillus subtilis | | Descriptor: | Cell division factor | | Authors: | Castellen, P, Sforca, M.L, Zeri, A.C.M, Gueiros-Filho, F.J. | | Deposit date: | 2014-07-16 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | FtsZ filament capping by MciZ, a developmental regulator of bacterial division.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1AZ4
 
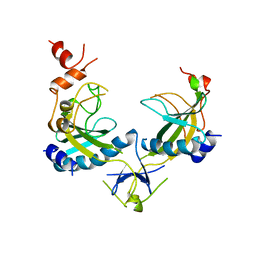 | | ECORV ENDONUCLEASE, UNLIGANDED, FORM B, T93A MUTANT | | Descriptor: | ECORV ENDONUCLEASE | | Authors: | Perona, J, Martin, A. | | Deposit date: | 1997-11-24 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational transitions and structural deformability of EcoRV endonuclease revealed by crystallographic analysis.
J.Mol.Biol., 273, 1997
|
|
1AZ3
 
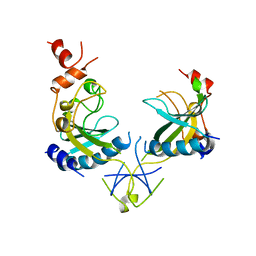 | | ECORV ENDONUCLEASE, UNLIGANDED, FORM B | | Descriptor: | ECORV ENDONUCLEASE | | Authors: | Perona, J, Martin, A. | | Deposit date: | 1997-11-24 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational transitions and structural deformability of EcoRV endonuclease revealed by crystallographic analysis.
J.Mol.Biol., 273, 1997
|
|
1PKR
 
 | |
1EAH
 
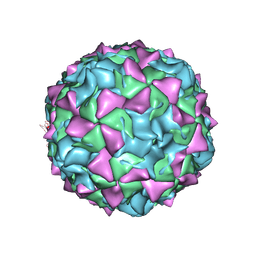 | | PV2L COMPLEXED WITH ANTIVIRAL AGENT SCH48973 | | Descriptor: | 1[2-CHLORO-4-METHOXY-PHENYL-OXYMETHYL]-4-[2,6-DICHLORO-PHENYL-OXYMETHYL]-BENZENE, MYRISTIC ACID, POLIOVIRUS TYPE 2 COAT PROTEINS VP1 TO VP4 | | Authors: | Lentz, K, Arnold, E. | | Deposit date: | 1997-07-22 | | Release date: | 1998-09-16 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of poliovirus type 2 Lansing complexed with antiviral agent SCH48973: comparison of the structural and biological properties of three poliovirus serotypes.
Structure, 5, 1997
|
|
1POV
 
 | |
2PLV
 
 | |
