4R8O
 
 | |
4R03
 
 | |
4RBO
 
 | |
1VKM
 
 | |
1VJ1
 
 | |
1VQR
 
 | |
1VR8
 
 | |
1VPZ
 
 | |
1VKH
 
 | |
1VK3
 
 | |
1VL4
 
 | |
1VJO
 
 | |
1VQ0
 
 | |
1VR3
 
 | |
1VRM
 
 | |
3SY6
 
 | |
3UFI
 
 | |
3UP6
 
 | |
3UWS
 
 | |
3U21
 
 | |
1AJ7
 
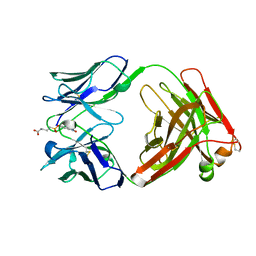 | | IMMUNOGLOBULIN 48G7 GERMLINE FAB ANTIBODY COMPLEXED WITH HAPTEN 5-(PARA-NITROPHENYL PHOSPHONATE)-PENTANOIC ACID. AFFINITY MATURATION OF AN ESTEROLYTIC ANTIBODY | | Descriptor: | 5-(PARA-NITROPHENYL PHOSPHONATE)-PENTANOIC ACID, IMMUNOGLOBULIN 48G7 FAB (HEAVY CHAIN), IMMUNOGLOBULIN 48G7 FAB (LIGHT CHAIN) | | Authors: | Wedemayer, G.J, Wang, L.H, Patten, P.A, Schultz, P.G, Stevens, R.C. | | Deposit date: | 1997-05-15 | | Release date: | 1997-11-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the evolution of an antibody combining site.
Science, 276, 1997
|
|
2B8N
 
 | |
2ETS
 
 | |
2F46
 
 | |
2FG0
 
 | |
