1U3W
 
 | |
3RT1
 
 | |
3SZB
 
 | |
3SZA
 
 | | Crystal structure of human ALDH3A1 - apo form | | Descriptor: | ACETATE ION, Aldehyde dehydrogenase, dimeric NADP-preferring, ... | | Authors: | Khanna, M, Hurley, T.D. | | Deposit date: | 2011-07-18 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Discovery of a novel class of covalent inhibitor for aldehyde dehydrogenases.
J.Biol.Chem., 286, 2011
|
|
3RSZ
 
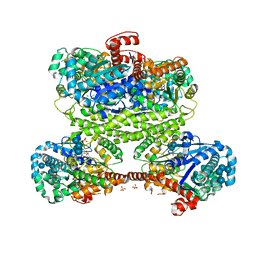 | | Maltodextran bound basal state conformation of yeast glycogen synthase isoform 2 | | Descriptor: | Glycogen [starch] synthase isoform 2, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Baskaran, S, Hurley, T.D. | | Deposit date: | 2011-05-02 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.009 Å) | | Cite: | Multiple Glycogen-binding Sites in Eukaryotic Glycogen Synthase Are Required for High Catalytic Efficiency toward Glycogen.
J.Biol.Chem., 286, 2011
|
|
4KQM
 
 | |
3O3C
 
 | | Glycogen synthase basal state UDP complex | | Descriptor: | Glycogen [starch] synthase isoform 2, SULFATE ION, URIDINE-5'-DIPHOSPHATE | | Authors: | Baskaran, S, Hurley, T.D. | | Deposit date: | 2010-07-23 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.512 Å) | | Cite: | Structural basis for glucose-6-phosphate activation of glycogen synthase.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1AG8
 
 | |
3NCH
 
 | |
1A4Z
 
 | |
3NAZ
 
 | | Basal state form of Yeast Glycogen Synthase | | Descriptor: | Glycogen [starch] synthase isoform 2, PEPTIDE, SULFATE ION | | Authors: | Baskaran, S, Hurley, T.D. | | Deposit date: | 2010-06-02 | | Release date: | 2010-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for glucose-6-phosphate activation of glycogen synthase.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3NB0
 
 | |
1M6H
 
 | | Human glutathione-dependent formaldehyde dehydrogenase | | Descriptor: | Glutathione-dependent formaldehyde dehydrogenase, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Sanghani, P.C, Robinson, H, Bosron, W.F, Hurley, T.D. | | Deposit date: | 2002-07-16 | | Release date: | 2002-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human glutathione-dependent formaldehyde dehydrogenase. Structures of apo, binary, and inhibitory ternary complexes.
Biochemistry, 41, 2002
|
|
1MA0
 
 | | Ternary complex of Human glutathione-dependent formaldehyde dehydrogenase with NAD+ and dodecanoic acid | | Descriptor: | Glutathione-dependent formaldehyde dehydrogenase, LAURIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Sanghani, P.C, Robinson, H, Bosron, W.F, Hurley, T.D. | | Deposit date: | 2002-07-30 | | Release date: | 2002-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human glutathione-dependent formaldehyde dehydrogenase. Structures of apo, binary, and inhibitory ternary complexes.
Biochemistry, 41, 2002
|
|
1M6W
 
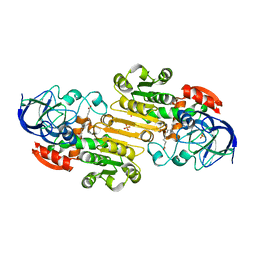 | | Binary complex of Human glutathione-dependent formaldehyde dehydrogenase and 12-Hydroxydodecanoic acid | | Descriptor: | 12-HYDROXYDODECANOIC ACID, Glutathione-dependent formaldehyde dehydrogenase, PHOSPHATE ION, ... | | Authors: | Sanghani, P.C, Robinson, H, Bosron, W.F, Hurley, T.D. | | Deposit date: | 2002-07-17 | | Release date: | 2002-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human glutathione-dependent formaldehyde dehydrogenase. Structures of apo, binary, and inhibitory ternary complexes.
Biochemistry, 41, 2002
|
|
1MC5
 
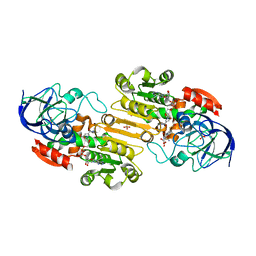 | | Ternary complex of Human glutathione-dependent formaldehyde dehydrogenase with S-(hydroxymethyl)glutathione and NADH | | Descriptor: | 2-AMINO-4-[1-CARBOXYMETHYL-CARBAMOYL)-2-HYDROXYMETHYLSULFANYL-ETHYLCARBAMOYL]-BUTYRIC ACID, Alcohol dehydrogenase class III chi chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Sanghani, P.C, Bosron, W.F, Hurley, T.D. | | Deposit date: | 2002-08-05 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human glutathione-dependent formaldehyde dehydrogenase.
Structural changes associated with Ternary Complex formation
Biochemistry, 41, 2002
|
|
1MP0
 
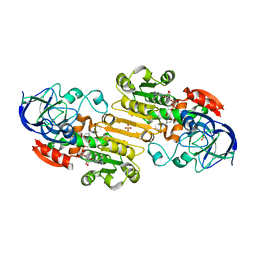 | | Binary Complex of Human Glutathione-Dependent Formaldehyde Dehydrogenase with NAD(H) | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Sanghani, P.C, Robinson, H, Hurley, T.D, Bosron, W.F. | | Deposit date: | 2002-09-10 | | Release date: | 2002-09-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-function relationships in human Class III alcohol dehydrogenase
(formaldehyde dehydrogenase)
Chem.Biol.Interact., 143, 2003
|
|
1LL2
 
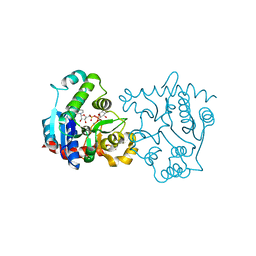 | | Crystal Structure of Rabbit Muscle Glycogenin Complexed with UDP-glucose and Manganese | | Descriptor: | GLYCOGENIN-1, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Gibbons, B.J, Roach, P.J, Hurley, T.D. | | Deposit date: | 2002-04-26 | | Release date: | 2002-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the autocatalytic initiator of glycogen biosynthesis, glycogenin.
J.Mol.Biol., 319, 2002
|
|
1LL3
 
 | |
1HSO
 
 | | HUMAN ALPHA ALCOHOL DEHYDROGENASE (ADH1A) | | Descriptor: | 4-IODOPYRAZOLE, CLASS I ALCOHOL DEHYDROGENASE 1, ALPHA SUBUNIT, ... | | Authors: | Niederhut, M.S, Gibbons, B.J, Perez-Miller, S, Hurley, T.D. | | Deposit date: | 2000-12-27 | | Release date: | 2001-01-10 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structures of the three human class I alcohol dehydrogenases.
Protein Sci., 10, 2001
|
|
1HT0
 
 | | HUMAN GAMMA-2 ALCOHOL DEHYDROGENSE | | Descriptor: | CLASS I ALCOHOL DEHYDROGENASE 3, GAMMA SUBUNIT, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Niederhut, M.S, Gibbons, B.J, Perez-Miller, S, Hurley, T.D. | | Deposit date: | 2000-12-27 | | Release date: | 2001-01-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structures of the three human class I alcohol dehydrogenases.
Protein Sci., 10, 2001
|
|
1LL0
 
 | |
1HSZ
 
 | | HUMAN BETA-1 ALCOHOL DEHYDROGENASE (ADH1B*1) | | Descriptor: | CLASS I ALCOHOL DEHYDROGENASE 1, BETA SUBUNIT, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Niederhut, M.S, Gibbons, B.J, Perez-Miller, S, Hurley, T.D. | | Deposit date: | 2000-12-27 | | Release date: | 2001-01-10 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structures of the three human class I alcohol dehydrogenases.
Protein Sci., 10, 2001
|
|
2ONN
 
 | |
2ONP
 
 | | Arg475Gln Mutant of Human Mitochondrial Aldehyde Dehydrogenase, complexed with NAD+ | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde dehydrogenase, GUANIDINE, ... | | Authors: | Larson, H.N, Hurley, T.D. | | Deposit date: | 2007-01-24 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional consequences of coenzyme binding to the inactive asian variant of mitochondrial aldehyde dehydrogenase: roles of residues 475 and 487.
J.Biol.Chem., 282, 2007
|
|
