2PWF
 
 | | Crystal structure of the MutB D200A mutant in complex with glucose | | Descriptor: | CALCIUM ION, Sucrose isomerase, beta-D-glucopyranose | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2007-05-11 | | Release date: | 2007-06-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Trehalulose synthase native and carbohydrate complexed structures provide insights into sucrose isomerization.
J.Biol.Chem., 61, 2007
|
|
2PWG
 
 | | Crystal Structure of the Trehalulose Synthase MutB From Pseudomonas Mesoacidophila MX-45 Complexed to the Inhibitor Castanospermine | | Descriptor: | CALCIUM ION, CASTANOSPERMINE, Sucrose isomerase | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2007-05-11 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trehalulose synthase native and carbohydrate complexed structures provide insights into sucrose isomerization.
J.Biol.Chem., 61, 2007
|
|
2PWE
 
 | | Crystal structure of the MutB E254Q mutant in complex with the substrate sucrose | | Descriptor: | CALCIUM ION, Sucrose isomerase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2007-05-11 | | Release date: | 2007-06-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Trehalulose synthase native and carbohydrate complexed structures provide insights into sucrose isomerization.
J.Biol.Chem., 282, 2007
|
|
2PWH
 
 | | Crystal structure of the trehalulose synthase MutB from Pseudomonas mesoacidophila MX-45 | | Descriptor: | CALCIUM ION, Sucrose isomerase | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2007-05-11 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Trehalulose synthase native and carbohydrate complexed structures provide insights into sucrose isomerization.
J.Biol.Chem., 61, 2007
|
|
2O4F
 
 | | Structure of a parallel-stranded guanine tetraplex crystallised with monovalent ions | | Descriptor: | 5'-D(*TP*GP*GP*GP*GP*T)-3', LITHIUM ION, SODIUM ION | | Authors: | Creze, C, Rinaldi, B, Haser, R, Bouvet, P, Gouet, P. | | Deposit date: | 2006-12-04 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a d(TGGGGT) quadruplex crystallized in the presence of Li+ ions.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2QPS
 
 | | "Sugar tongs" mutant Y380A in complex with acarbose | | Descriptor: | Alpha-amylase type A isozyme, CALCIUM ION | | Authors: | Aghajari, N, Jensen, M.H, Tranier, S, Haser, R. | | Deposit date: | 2007-07-25 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 'pair of sugar tongs' site on the non-catalytic domain C of barley alpha-amylase participates in substrate binding and activity
Febs J., 274, 2007
|
|
2QNO
 
 | | Crystal Structure of the Mutant E55Q of the Cellulase CEL48F in Complex with a Thio-Oligosaccharide | | Descriptor: | 4-thio-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose, CALCIUM ION, Endoglucanase F | | Authors: | Parsiegla, G, Haser, R. | | Deposit date: | 2007-07-19 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of mutants of cellulase Cel48F of Clostridium cellulolyticum in complex with long hemithiocellooligosaccharides give rise to a new view of the substrate pathway during processive action
J.Mol.Biol., 375, 2008
|
|
3GBE
 
 | | Crystal structure of the isomaltulose synthase SmuA from Protaminobacter rubrum in complex with the inhibitor deoxynojirimycin | | Descriptor: | 1,2-ETHANEDIOL, 1-DEOXYNOJIRIMYCIN, CITRATE ANION, ... | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2009-02-19 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural determinants of product specificity of sucrose isomerases
Febs Lett., 583, 2009
|
|
3GBD
 
 | | Crystal structure of the isomaltulose synthase SmuA from Protaminobacter rubrum | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Sucrose isomerase SmuA from Protaminobacter rubrum | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2009-02-19 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural determinants of product specificity of sucrose isomerases
Febs Lett., 583, 2009
|
|
4FNR
 
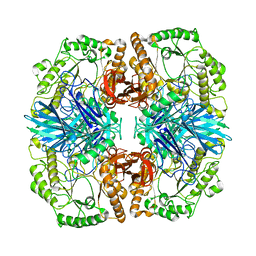 | | Crystal structure of GH36 alpha-galactosidase AgaA from Geobacillus stearothermophilus | | Descriptor: | Alpha-galactosidase AgaA | | Authors: | Merceron, R, Foucault, M, Haser, R, Mattes, R, Watzlawick, H, Gouet, P. | | Deposit date: | 2012-06-20 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The molecular mechanism of the thermostable alpha-galactosidases AgaA and AgaB explained by X-ray crystallography and mutational studies
J.Biol.Chem., 287, 2012
|
|
4FNS
 
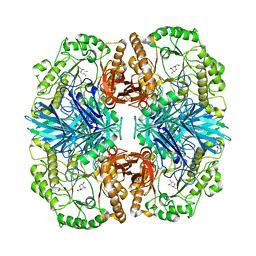 | | Crystal structure of GH36 alpha-galactosidase AgaA A355E from Geobacillus stearothermophilus in complex with 1-deoxygalactonojirimycin | | Descriptor: | (2R,3S,4R,5S)-2-(hydroxymethyl)piperidine-3,4,5-triol, 1,2-ETHANEDIOL, Alpha-galactosidase AgaA, ... | | Authors: | Merceron, R, Foucault, M, Haser, R, Mattes, R, Watzlawick, H, Gouet, P. | | Deposit date: | 2012-06-20 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The molecular mechanism of the thermostable alpha-galactosidases AgaA and AgaB explained by X-ray crystallography and mutational studies
J.Biol.Chem., 287, 2012
|
|
4GIA
 
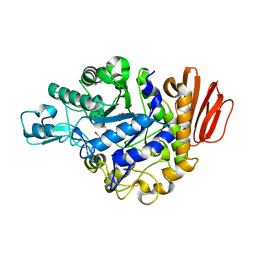 | | Crystal structure of the MUTB F164L mutant from crystals soaked with isomaltulose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Lipski, A, Haser, R, Aghajari, N. | | Deposit date: | 2012-08-08 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Mutations inducing an active-site aperture in Rhizobium sp. sucrose isomerase confer hydrolytic activity
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GI9
 
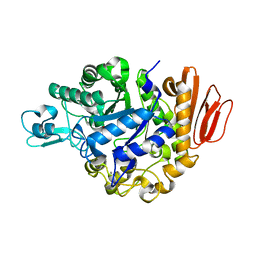 | | Crystal structure of the MUTB F164L mutant from crystals soaked with Trehalulose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Lipski, A, Haser, R, Aghajari, N. | | Deposit date: | 2012-08-08 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mutations inducing an active-site aperture in Rhizobium sp. sucrose isomerase confer hydrolytic activity
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GI8
 
 | | Crystal structure of the MUTB F164L mutant from crystals soaked with the substrate sucrose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Lipski, A, Haser, R, Aghajari, N. | | Deposit date: | 2012-08-08 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mutations inducing an active-site aperture in Rhizobium sp. sucrose isomerase confer hydrolytic activity
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GIN
 
 | | Crystal structure of the MUTB R284C mutant from crystals soaked with the inhibitor deoxynojirimycin | | Descriptor: | CALCIUM ION, GLYCEROL, Sucrose isomerase | | Authors: | Lipski, A, Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2012-08-08 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutations inducing an active-site aperture in Rhizobium sp. sucrose isomerase confer hydrolytic activity
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GI6
 
 | | Crystal structure of the MUTB F164L mutant in complex with glucose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Lipski, A, Haser, R, Aghajari, N. | | Deposit date: | 2012-08-08 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mutations inducing an active-site aperture in Rhizobium sp. sucrose isomerase confer hydrolytic activity
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4H8H
 
 | | MUTB inactive double mutant E254Q-D415N | | Descriptor: | CALCIUM ION, GLYCEROL, SULFATE ION, ... | | Authors: | Lipski, A, Haser, R, Aghajari, N. | | Deposit date: | 2012-09-22 | | Release date: | 2013-09-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into product binding in sucrose isomerases from crystal structures of MutB from Rhizobium sp.
To be Published
|
|
4H8V
 
 | |
4GO8
 
 | | Crystal Structure of the TREHALULOSE SYNTHASE MUTB, MUTANT A258V, in complex with TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Sucrose isomerase | | Authors: | Lipski, A, Haser, R, Aghajari, N. | | Deposit date: | 2012-08-19 | | Release date: | 2013-08-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of MUTB A258V mutant in complex with TRIS
To be Published, 2013
|
|
4H2C
 
 | | Trehalulose synthase MutB R284C mutant | | Descriptor: | CALCIUM ION, GLYCEROL, Sucrose isomerase | | Authors: | Lipski, A, Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2012-09-12 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mutations inducing an active-site aperture in Rhizobium sp. sucrose isomerase confer hydrolytic activity
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GO9
 
 | | CRYSTAL STRUCTURE of the TREHALULOSE SYNTHASE MUTANT, MUTB D415N, in COMPLEX with TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Sucrose isomerase | | Authors: | Lipski, A, Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2012-08-19 | | Release date: | 2013-08-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | CRYSTAL STRUCTURE of the TREHALULOSE SYNTHASE MUTB, MUTANT D415N, in COMPLEX with TRIS
To be Published, 2013
|
|
4HA1
 
 | |
4H7V
 
 | |
4H8U
 
 | |
4HHM
 
 | | Crystal structure of a mutant, G219A, of Glucose Isomerase from Streptomyces sp. SK | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, Xylose isomerase | | Authors: | Ben Hlima, H, Riguet, J, Haser, R, Aghajari, N. | | Deposit date: | 2012-10-10 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification of critical residues for the activity and thermostability of Streptomyces sp. SK glucose isomerase.
Appl.Microbiol.Biotechnol., 97, 2013
|
|
