9AYX
 
 | |
6XRJ
 
 | |
6VTU
 
 | | DH717.1 Fab monomer in complex with man9 glycan | | Descriptor: | DH717.1 heavy chain, DH717.1 light chain, GLYCEROL, ... | | Authors: | Fera, D, Bronkema, N. | | Deposit date: | 2020-02-13 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
8V0X
 
 | |
8UKF
 
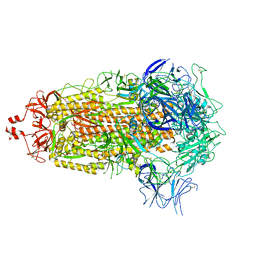 | | SARS-CoV-2 Omicron-EG.5 3-RBD down Spike Protein Trimer consensus (S-GSAS-Omicron-EG.5) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-10-12 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition
Mol.Cell, 84, 2024
|
|
8V0N
 
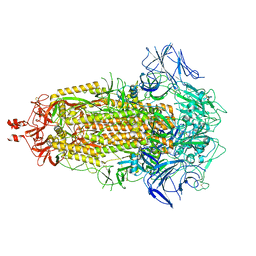 | | SARS-CoV-2 Omicron-XBB.1.16 3-RBD-down Spike Protein Trimer 3 (S-RRAR-Omicron-XBB.1.16) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
8V0W
 
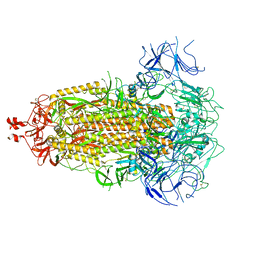 | | SARS-CoV-2 Omicron-EG.5 3-RBD down Spike Protein Trimer 2 (S-GSAS-Omicron-EG.5) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
8V0R
 
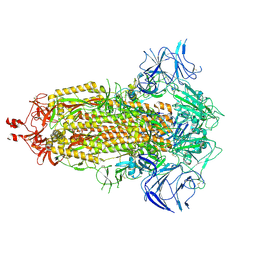 | | SARS-CoV-2 Omicron-XBB.1.5 3-RBD down Spike Protein Trimer 1 (S-GSAS-Omicron-XBB.1.5) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
8V0U
 
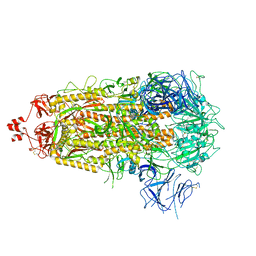 | | SARS-CoV-2 Omicron-XBB.1.5 3-RBD down Spike Protein Trimer 4 (S-GSAS-Omicron-XBB.1.5) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
8UKD
 
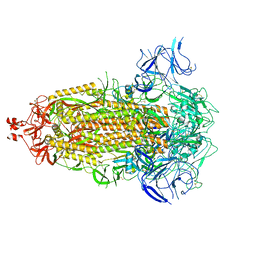 | |
8V0Q
 
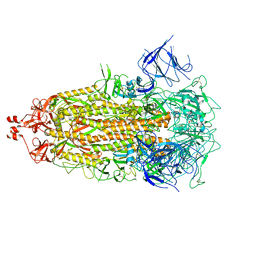 | | SARS-CoV-2 Omicron-XBB.1.16 3-RBD down Spike Protein Trimer 3 (S-GSAS-Omicron-XBB.1.16) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
8V0P
 
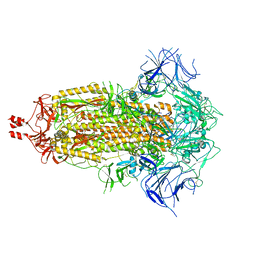 | | SARS-CoV-2 Omicron-XBB.1.16 3-RBD down Spike Protein Trimer 2 (S-GSAS-Omicron-XBB.1.16) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
8V0O
 
 | | SARS-CoV-2 Omicron-XBB.1.16 3-RBD down Spike Protein Trimer 1 (S-GSAS-Omicron-XBB.1.16) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
8UK1
 
 | | SARS-CoV-2 Omicron-XBB.1.16 3-RBD-down Spike Protein Trimer consensus (S-RRAR-Omicron-XBB.1.16) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-10-11 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
8V0T
 
 | |
8V0M
 
 | | SARS-CoV-2 Omicron-XBB.1.16 3-RBD-down Spike Protein Trimer 2 (S-RRAR-Omicron-XBB.1.16) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
8V0L
 
 | | SARS-CoV-2 Omicron-XBB.1.16 3-RBD-down Spike Protein Trimer 1 (S-RRAR-Omicron-XBB.1.16) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
8V0V
 
 | | SARS-CoV-2 Omicron-EG.5 3-RBD down Spike Protein Trimer 1 (S-GSAS-Omicron-EG.5) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
8V0S
 
 | | SARS-CoV-2 Omicron-XBB.1.5 3-RBD down Spike Protein Trimer 2 (S-GSAS-Omicron-XBB.1.5) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
8UIR
 
 | | SARS-CoV-2 Omicron-XBB.1.16 3-RBD down Spike Protein Trimer consensus (S-GSAS-Omicron-XBB.1.16) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-10-10 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
7L06
 
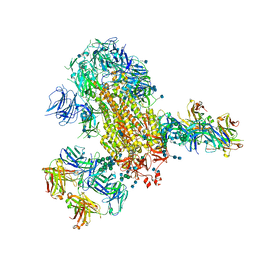 | | Cryo-EM structure of SARS-CoV-2 2P S ectodomain bound to two copies of domain-swapped antibody 2G12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2G12 heavy chain, ... | | Authors: | Manne, K, Henderson, R, Acharya, P. | | Deposit date: | 2020-12-11 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
7L09
 
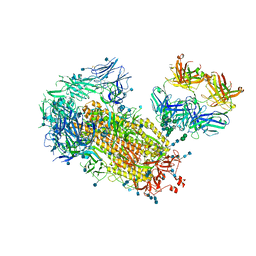 | | Cryo-EM structure of SARS-CoV-2 2P S ectodomain bound domain-swapped antibody 2G12 from masked 3D refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2G12 heavy chain, ... | | Authors: | Manne, K, Henderson, R, Acharya, P. | | Deposit date: | 2020-12-11 | | Release date: | 2020-12-30 | | Last modified: | 2021-06-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
7L02
 
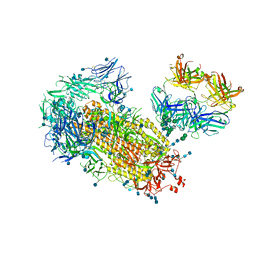 | | Cryo-EM structure of SARS-CoV-2 2P S ectodomain bound to one copy of domain-swapped antibody 2G12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2G12 heavy chain, ... | | Authors: | Manne, K, Henderson, R, Acharya, P. | | Deposit date: | 2020-12-10 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
7L6M
 
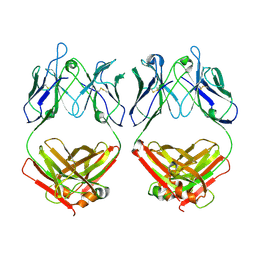 | |
7LUA
 
 | | Cryo-EM structure of DH898.1 Fab-dimer bound near the CD4 binding site of HIV-1 Env CH848 SOSIP trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848 SOSIP gp120, ... | | Authors: | Manne, K, Edwards, R.J, Acharya, P. | | Deposit date: | 2021-02-21 | | Release date: | 2021-03-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
