3IK4
 
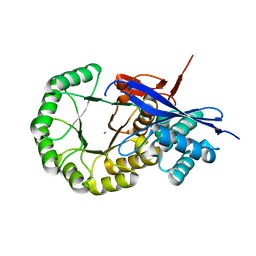 | | CRYSTAL STRUCTURE OF mandelate racemase/muconate lactonizing protein from Herpetosiphon aurantiacus | | Descriptor: | GLYCEROL, Mandelate racemase/muconate lactonizing protein, POTASSIUM ION | | Authors: | Patskovsky, Y, Toro, R, Dickey, M, Iizuka, M, Sauder, J.M, Gerlt, J.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-05 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3IJQ
 
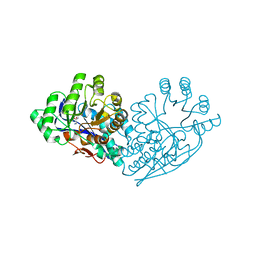 | | Structure of dipeptide epimerase from Bacteroides thetaiotaomicron complexed with L-Ala-D-Glu; productive substrate binding. | | Descriptor: | ALANINE, D-GLUTAMIC ACID, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Lukk, T, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-08-04 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3IJL
 
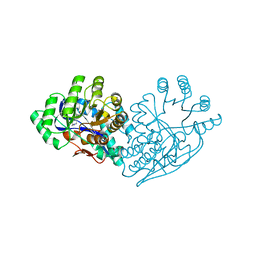 | | Structure of dipeptide epimerase from Bacteroides thetaiotaomicron complexed with L-Pro-D-Glu; nonproductive substrate binding. | | Descriptor: | D-GLUTAMIC ACID, MAGNESIUM ION, Muconate cycloisomerase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Lukk, T, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-08-04 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3JZU
 
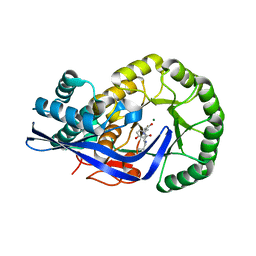 | | Crystal structure of Dipeptide Epimerase from Enterococcus faecalis V583 complexed with Mg and dipeptide L-Leu-L-Tyr | | Descriptor: | Dipeptide Epimerase, LEUCINE, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Sakai, A, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-09-24 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3KUM
 
 | | Crystal structure of Dipeptide Epimerase from Enterococcus faecalis V583 complexed with Mg and dipeptide L-Arg-L-Tyr | | Descriptor: | ARGININE, Dipeptide Epimerase, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Sakai, A, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-11-27 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3JVA
 
 | | Crystal structure of Dipeptide Epimerase from Enterococcus faecalis V583 | | Descriptor: | Dipeptide Epimerase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Sakai, A, Imker, H, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-09-16 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3JW7
 
 | | Crystal structure of Dipeptide Epimerase from Enterococcus faecalis V583 complexed with Mg and dipeptide L-Ile-L-Tyr | | Descriptor: | Dipeptide Epimerase, GLYCEROL, ISOLEUCINE, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Sakai, A, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-09-18 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3K1G
 
 | | Crystal structure of Dipeptide Epimerase from Enterococcus faecalis V583 complexed with Mg and dipeptide L-Ser-L-Tyr | | Descriptor: | Dipeptide Epimerase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Sakai, A, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-09-27 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5HFK
 
 | | CRYSTAL STRUCTURE OF A GLUTATHIONE S-TRANSFERASE PROTEIN FROM ESCHERICHIA COLI OCh 157:H7 STR. SAKAI (ECs3186, TARGET EFI-507414) WITH BOUND GLUTATHIONE | | Descriptor: | Disulfide-bond oxidoreductase YfcG, GLUTATHIONE | | Authors: | Himmel, D.M, Toro, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-01-07 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | CRYSTAL STRUCTURE OF A GLUTATHIONE S-TRANSFERASE PROTEIN FROM ESCHERICHIA COLI OCh 157:H7 STR. SAKAI (ECs3186, TARGET EFI-507414) WITH BOUND GLUTATHIONE
TO BE PUBLISHED
|
|
5HSG
 
 | | Crystal structure of an ABC transporter Solute Binding Protein from Klebsiella pneumoniae (KPN_01730, target EFI-511059), APO open structure | | Descriptor: | MAGNESIUM ION, Putative ABC transporter, nucleotide binding/ATPase protein | | Authors: | Roth, Y, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2016-01-25 | | Release date: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of an ABC transporter Solute Binding Protein from Klebsiella pneumoniae (KPN_01730, target EFI-511059), APO open structure
To be published
|
|
5HKO
 
 | | Crystal structure of ABC transporter Solute Binding Protein MSMEG_3598 from Mycobacterium smegmatis str. MC2 155, target EFI-510969, in complex with L-sorbitol | | Descriptor: | ABC transporter, carbohydrate uptake transporter-2 (CUT2) family, periplasmic sugar-binding protein, ... | | Authors: | Roth, Y, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2016-01-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of ABC transporter Solute Binding Protein MSMEG_3598 from Mycobacterium smegmatis str. MC2 155, target EFI-510969, in complex with L-sorbitol
To be published
|
|
5IAI
 
 | | Crystal structure of ABC transporter Solute Binding Protein Arad_9887 from Agrobacterium radiobacter K84, target EFI-510945 in complex with Ribitol | | Descriptor: | D-ribitol, GLYCEROL, Sugar ABC transporter | | Authors: | Vetting, M.W, Bonanno, J.B, Al Obaidi, N.F, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2016-02-21 | | Release date: | 2016-03-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of ABC transporter Solute Binding Protein Arad_9887 from Agrobacterium radiobacter K84, target EFI-510945 in complex with Ribitol
To be published
|
|
5IM2
 
 | | Crystal structure of a TRAP solute binding protein from Rhodoferax ferrireducens T118 (Rfer_2570, TARGET EFI-510210) in complex with copurified benzoate | | Descriptor: | BENZOIC ACID, Twin-arginine translocation pathway signal | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2016-03-05 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a TRAP solute binding protein from Rhodoferax ferrireducens T118 (Rfer_2570, TARGET EFI-510210) in complex with copurified benzoate
To be published
|
|
5IBQ
 
 | | Crystal structure of an ABC solute binding protein from Rhizobium etli CFN 42 (RHE_PF00037,TARGET EFI-511357) in complex with alpha-D-apiose | | Descriptor: | 3-C-(hydroxylmethyl)-alpha-D-erythrofuranose, CALCIUM ION, Probable ribose ABC transporter, ... | | Authors: | Vetting, M.W, Carter, M.S, Al Obaidi, N.F, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2016-02-22 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of an ABC solute binding protein from Rhizobium etli CFN 42 (RHE_PF00037,TARGET EFI-511357) in complex with alpha-D-apiose
To be published
|
|
5HQJ
 
 | | Crystal structure of ABC transporter Solute Binding Protein B1G1H7 from Burkholderia graminis C4D1M, target EFI-511179, in complex with D-arabinose | | Descriptor: | CHLORIDE ION, Periplasmic binding protein/LacI transcriptional regulator, alpha-D-arabinopyranose | | Authors: | Roth, Y, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2016-01-21 | | Release date: | 2016-03-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of ABC transporter Solute Binding Protein B1G1H7 from Burkholderia graminis C4D1M, target EFI-511179, in complex with D-arabinose
To be published
|
|
4YZZ
 
 | | Crystal structure of a TRAP transporter solute binding protein (IPR025997) from Bordetella bronchiseptica RB50 (BB0280, TARGET EFI-500035) mixed occupancy dimer, copurified calcium and picolinate bound active site versus apo site | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-25 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of a TRAP transporter solute binding protein (IPR025997) from Bordetella bronchiseptica RB50 (BB0280, TARGET EFI-500035) mixed occupancy dimer, copurified calcium and picolinate bound active site versus apo site
To be published
|
|
4ZJP
 
 | | Structure of an ABC-Transporter Solute Binding Protein (SBP_IPR025997) from Actinobacillus Succinogenes (Asuc_0197, TARGET EFI-511067) with bound beta-D-ribopyranose | | Descriptor: | 1,2-ETHANEDIOL, Monosaccharide-transporting ATPase, beta-D-ribopyranose | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Glenn, A.S, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure of an ABC-Transporter Solute Binding Protein (SBP_IPR025997) from Actinobacillus Succinogenes (Asuc_0197, TARGET EFI-511067) with bound beta-D-ribopyranose
To be published
|
|
5BR1
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS S4 (Avi_5305, TARGET EFI-511224) WITH BOUND ALPHA-D-GALACTOSAMINE | | Descriptor: | 2-amino-2-deoxy-alpha-D-galactopyranose, ABC transporter, binding protein | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-05-29 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of an ABC transporter solute-binding protein specific for the amino sugars glucosamine and galactosamine.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4Z0N
 
 | | Crystal Structure of a Periplasmic Solute binding protein (IPR025997) from Streptobacillus moniliformis DSM-12112 (Smon_0317, TARGET EFI-511281) with bound D-Galactose | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-26 | | Release date: | 2015-04-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal Structure of a Periplasmic Solute binding protein (IPR025997) from Streptobacillus moniliformis DSM-12112 (Smon_0317, TARGET EFI-511281) with bound D-Galactose
To be published
|
|
4ZQ8
 
 | | Crystal structure of a terpene synthase from Streptomyces lydicus, target EFI-540129 | | Descriptor: | Isoprenoid Synthase | | Authors: | Toro, R, Bhosle, R, Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Poulter, C.D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-05-08 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a terpene synthase from Streptomyces lydicus, target EFI-540129
To be published
|
|
5CI5
 
 | | Crystal Structure of an ABC transporter Solute Binding Protein from Thermotoga Lettingae TMO (Tlet_1705, TARGET EFI-510544) bound with alpha-D-Tagatose | | Descriptor: | 1,2-ETHANEDIOL, Extracellular solute-binding protein family 1, PENTAETHYLENE GLYCOL, ... | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-07-11 | | Release date: | 2015-07-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of an ABC transporter Solute Binding Protein from Thermotoga Lettingae TMO (Tlet_1705, TARGET EFI-510544) bound with alpha-D-Tagatose
To be published
|
|
5CM6
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM PSEUDOALTEROMONAS ATLANTICA T6c(Patl_2292, TARGET EFI-510180) WITH BOUND SODIUM AND PYRUVATE | | Descriptor: | PYRUVIC ACID, SODIUM ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-07-16 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM PSEUDOALTEROMONAS ATLANTICA T6c(Patl_2292, TARGET EFI-510180) WITH BOUND SODIUM AND PYRUVATE
To be published
|
|
5BRA
 
 | | Crystal Structure of a putative Periplasmic Solute binding protein (IPR025997) from Ochrobactrum Anthropi ATCC49188 (Oant_2843, TARGET EFI-511085) | | Descriptor: | Putative periplasmic binding protein with substrate ribose | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-05-30 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.971 Å) | | Cite: | Crystal Structure of a putative Periplasmic Solute binding protein (IPR025997) from Ochrobactrum Anthropi ATCC49188(Oant_2843, TARGET EFI-511085)
To be published
|
|
1Q6Q
 
 | | Structure of 3-keto-L-gulonate 6-phosphate decarboxylase with bound xylitol 5-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-XYLITOL 5-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | Deposit date: | 2003-08-13 | | Release date: | 2003-10-28 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Structural Evidence for a 1,2-Enediolate Intermediate in the Reaction Catalyzed by 3-Keto-l-Gulonate 6-Phosphate Decarboxylase, a Member of the Orotidine 5'-Monophosphate Decarboxylase Suprafamily
Biochemistry, 42, 2003
|
|
1Q6O
 
 | | Structure of 3-keto-L-gulonate 6-phosphate decarboxylase with bound L-gulonaet 6-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-GULURONIC ACID 6-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | Deposit date: | 2003-08-13 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.202 Å) | | Cite: | Structural Evidence for a 1,2-Enediolate Intermediate in the Reaction Catalyzed by 3-Keto-l-Gulonate 6-Phosphate Decarboxylase, a Member of the Orotidine 5'-Monophosphate Decarboxylase Suprafamily
Biochemistry, 42, 2003
|
|
