3N7Q
 
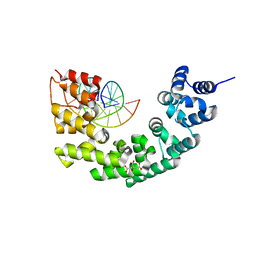 | | Crystal structure of human mitochondrial mTERF fragment (aa 99-399) in complex with a 12-mer DNA encompassing the tRNALeu(UUR) binding sequence | | Descriptor: | CITRIC ACID, DNA (5'-D(*CP*CP*GP*GP*GP*CP*TP*CP*TP*GP*CP*C)-3'), DNA (5'-D(*GP*GP*CP*AP*GP*AP*GP*CP*CP*CP*GP*G)-3'), ... | | Authors: | Jimenez-Menendez, N, Fernandez-Millan, P, Rubio-Cosials, A, Arnan, C, Montoya, J, Jacobs, H.T, Bernado, P, Coll, M, Uson, I, Sola, M. | | Deposit date: | 2010-05-27 | | Release date: | 2010-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human mitochondrial mTERF wraps around DNA through a left-handed superhelical tandem repeat.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4ICW
 
 | |
1NQ6
 
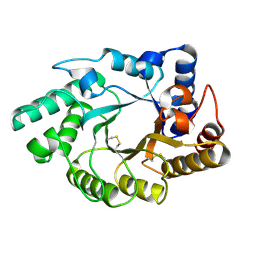 | | Crystal Structure of the catalytic domain of xylanase A from Streptomyces halstedii JM8 | | Descriptor: | MAGNESIUM ION, Xys1 | | Authors: | Canals, A, Vega, M.C, Gomis-Ruth, F.X, Santamaria, R.I, Coll, M. | | Deposit date: | 2003-01-21 | | Release date: | 2004-01-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of xylanase Xys1delta from Streptomyces halstedii.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4ICX
 
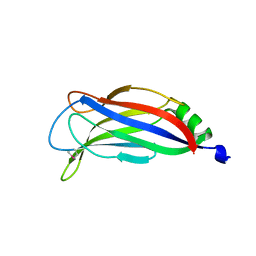 | |
4LDU
 
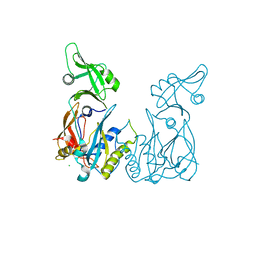 | | Crystal structure of the DNA binding domain of Arabidopsis thaliana auxin response factor 5 | | Descriptor: | Auxin response factor 5, CHLORIDE ION | | Authors: | Boer, D.R, Freire-Rios, A, van den Berg, W.M.A, Weijers, D, Coll, M. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for DNA Binding Specificity by the Auxin-Dependent ARF Transcription Factors.
Cell(Cambridge,Mass.), 156, 2014
|
|
4LDW
 
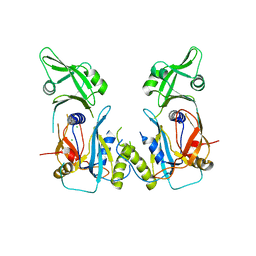 | |
1DZA
 
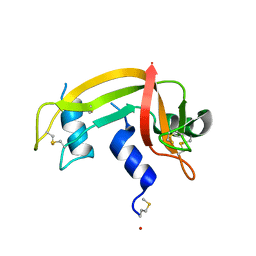 | | 3-D structure of a HP-RNase | | Descriptor: | RIBONUCLEASE 1 | | Authors: | Pous, J, Canals, A, Terzyan, S.S, Guasch, A, Benito, A, Ribo, M, Vilanova, M, Coll, M. | | Deposit date: | 2000-02-21 | | Release date: | 2001-02-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Three-Dimensional Structure of a Human Pancreatic Ribonuclease Variant, a Step Forward in the Design of Cytotoxic Ribonucleases
J.Mol.Biol., 303, 2000
|
|
1E6M
 
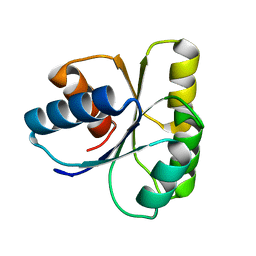 | | TWO-COMPONENT SIGNAL TRANSDUCTION SYSTEM D57A MUTANT OF CHEY | | Descriptor: | CHEMOTAXIS PROTEIN CHEY | | Authors: | Sola, M, Lopez-Hernandez, E, Cronet, P, Lacroix, E, Serrano, L, Coll, M, Parraga, A. | | Deposit date: | 2000-08-18 | | Release date: | 2000-09-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Towards understanding a molecular switch mechanism: thermodynamic and crystallographic studies of the signal transduction protein CheY.
J.Mol.Biol., 303, 2000
|
|
1E6L
 
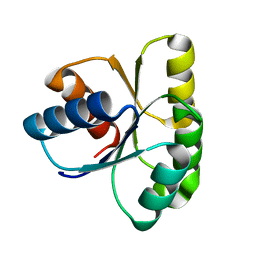 | | Two-component signal transduction system D13A mutant of CheY | | Descriptor: | Chemotaxis protein CheY | | Authors: | Sola, M, Lopez-Hernandez, E, Cronet, P, Lacroix, E, Serrano, L, Coll, M, Parraga, A. | | Deposit date: | 2000-08-18 | | Release date: | 2001-03-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Towards understanding a molecular switch mechanism: thermodynamic and crystallographic studies of the signal transduction protein CheY.
J.Mol.Biol., 303, 2000
|
|
1E21
 
 | | Ribonuclease 1 des1-7 Crystal Structure at 1.9A | | Descriptor: | RIBONUCLEASE 1 | | Authors: | Pous, J, Mallorqui-Fernandez, G, Peracaula, R, Terzyan, S.S, Futami, J, Tada, H, Yamada, H, Seno, M, De Llorens, R, Gomis-Ruth, F.X, Coll, M. | | Deposit date: | 2000-05-15 | | Release date: | 2001-05-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-Dimensional Crystal Structure of Human Rnase 1Dn7 at 1.9A Resolution
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1DYT
 
 | | X-ray crystal structure of ECP (RNase 3) at 1.75 A | | Descriptor: | CITRIC ACID, EOSINOPHIL CATIONIC PROTEIN, FE (III) ION | | Authors: | Mallorqui-Fernandez, G, Pous, J, Peracaula, R, Maeda, T, Tada, H, Yamada, H, Seno, M, De Llorens, R, Gomis-Rueth, F.X, Coll, M. | | Deposit date: | 2000-02-08 | | Release date: | 2001-02-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Three-Dimensional Crystal Structure of Human Eosinophil Cationic Protein (Rnase 3) at 1.75 A Resolution.
J.Mol.Biol., 300, 2000
|
|
1OKR
 
 | | Three-dimensional structure of S.aureus methicillin-resistance regulating transcriptional repressor MecI. | | Descriptor: | CHLORIDE ION, GLYCEROL, METHICILLIN RESISTANCE REGULATORY PROTEIN MECI | | Authors: | Garcia-Castellanos, R, Marrero, A, Mallorqui-Fernandez, G, Potempa, J, Coll, M, Gomis-Ruth, F.X. | | Deposit date: | 2003-07-28 | | Release date: | 2003-10-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-Dimensional Structure of Meci: Molecular Basis for Transcriptional Regulation of Staphylococcal Methicillin Resistance
J.Biol.Chem., 278, 2003
|
|
1E6K
 
 | | Two-component signal transduction system D12A mutant of CheY | | Descriptor: | Chemotaxis protein CheY | | Authors: | Sola, M, Lopez-Hernandez, E, Cronet, P, Lacroix, E, Serrano, L, Coll, M, Parraga, A. | | Deposit date: | 2000-08-18 | | Release date: | 2001-03-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Towards understanding a molecular switch mechanism: thermodynamic and crystallographic studies of the signal transduction protein CheY.
J.Mol.Biol., 303, 2000
|
|
3L0O
 
 | | Structure of RNA-free Rho transcription termination factor from Thermotoga maritima | | Descriptor: | SODIUM ION, SULFATE ION, Transcription termination factor rho, ... | | Authors: | Canals, A, Uson, I, Coll, M. | | Deposit date: | 2009-12-10 | | Release date: | 2010-05-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Structure of RNA-Free Rho Termination Factor Indicates a Dynamic Mechanism of Transcript Capture
J.Mol.Biol., 400, 2010
|
|
1CHN
 
 | |
1E9S
 
 | | Bacterial conjugative coupling protein TrwBdeltaN70. Unbound monoclinic form. | | Descriptor: | CONJUGAL TRANSFER PROTEIN TRWB | | Authors: | Gomis-Rueth, F.X, Moncalian, G, Cabezon, E, de la Cruz, F, Coll, M. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Bacterial Conjugation Protein Trwb Resembles Ring Helicases and F1-ATPase
Nature, 409, 2001
|
|
1D13
 
 | | MOLECULAR STRUCTURE OF AN A-DNA DECAMER D(ACCGGCCGGT) | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*GP*CP*CP*GP*GP*T)-3') | | Authors: | Frederick, C.A, Quigley, G.J, Teng, M.-K, Coll, M, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Wang, A.H.-J. | | Deposit date: | 1989-10-20 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular structure of an A-DNA decamer d(ACCGGCCGGT).
Eur.J.Biochem., 181, 1989
|
|
1E9R
 
 | | Bacterial conjugative coupling protein TrwBdeltaN70. Trigonal form in complex with sulphate. | | Descriptor: | CONJUGAL TRANSFER PROTEIN TRWB, SULFATE ION | | Authors: | Gomis-Rueth, F.X, Moncalian, G, Cabezon, E, de la Cruz, F, Coll, M. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Bacterial Conjugation Protein Trwb Resembles Ring Helicases and F1-ATPase
Nature, 409, 2001
|
|
1GLQ
 
 | |
2JB9
 
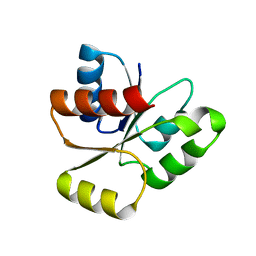 | | PhoB response regulator receiver domain constitutively-active double mutant D10A and D53E. | | Descriptor: | PHOSPHATE REGULON TRANSCRIPTIONAL REGULATORY PROTEIN PHOB | | Authors: | Ferrer-Orta, C, Arribas-Bosacoma, R, Kim, S.-K, Blanco, A.G, Pereira, P.J.B, Gomis-Ruth, F.X, Wanner, B.L, Coll, M, Sola, M. | | Deposit date: | 2006-12-05 | | Release date: | 2007-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The X-Ray Crystal Structures of Two Constitutively Active Mutants of the E. Coli Phob Receiver Domain Give Insights Into Activation
J.Mol.Biol., 366, 2007
|
|
2JBA
 
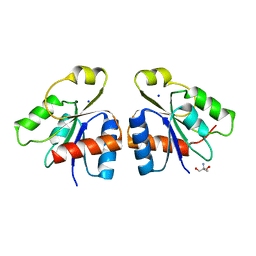 | | PhoB response regulator receiver domain constitutively-active double mutant D53A and Y102C. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE REGULON TRANSCRIPTIONAL REGULATORY PROTEIN PHOB, SODIUM ION | | Authors: | Arribas-Bosacoma, R, Ferrer-Orta, C, Kim, S.-K, Blanco, A.G, Pereira, P.J.B, Gomis-Ruth, F.X, Wanner, B.L, Coll, M, Sola, M. | | Deposit date: | 2006-12-05 | | Release date: | 2007-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The X-Ray Crystal Structures of Two Constitutively Active Mutants of the Escherichia Coli Phob Receiver Domain Give Insights Into Activation.
J.Mol.Biol., 366, 2007
|
|
2WYL
 
 | | Apo structure of a metallo-b-lactamase | | Descriptor: | FORMYL GROUP, GLYCEROL, L-ASCORBATE-6-PHOSPHATE LACTONASE ULAG | | Authors: | Garces, F, Fernandez, F.J, Penya-Soler, E, Aguilar, J, Baldoma, L, Coll, M, Badia, J, Vega, M.C. | | Deposit date: | 2009-11-16 | | Release date: | 2010-04-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Molecular Architecture of the Mn(2+)Dependent Lactonase Ulag Reveals an Rnase-Like Metallo-Beta-Lactamase Fold and a Novel Quaternary Structure.
J.Mol.Biol., 398, 2010
|
|
2WYM
 
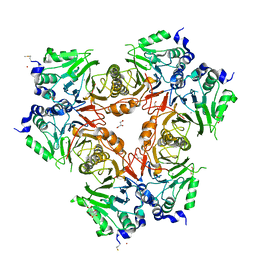 | | Structure of a metallo-b-lactamase | | Descriptor: | CITRATE ANION, GLYCEROL, L-ASCORBATE-6-PHOSPHATE LACTONASE ULAG, ... | | Authors: | Garces, F, Fernandez, F.J, Penya-Soler, E, Aguilar, J, Baldoma, L, Coll, M, Badia, J, Vega, M.C. | | Deposit date: | 2009-11-16 | | Release date: | 2010-04-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Architecture of the Mn(2+)Dependent Lactonase Ulag Reveals an Rnase-Like Metallo-Beta-Lactamase Fold and a Novel Quaternary Structure.
J.Mol.Biol., 398, 2010
|
|
1VTE
 
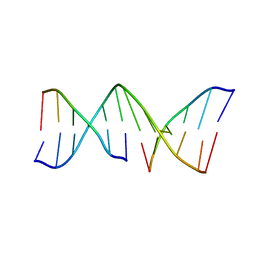 | | MOLECULAR STRUCTURE OF NICKED DNA. MODEL A4 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*AP*AP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*TP*T)-3'), DNA (5'-D(*TP*TP*CP*GP*CP*G)-3') | | Authors: | Aymani, J, Coll, M, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J, Rich, A. | | Deposit date: | 1990-05-21 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular structure of nicked DNA: a substrate for DNA repair enzymes.
Proc. Natl. Acad. Sci. U.S.A., 87, 1990
|
|
1YMV
 
 | |
