5NJT
 
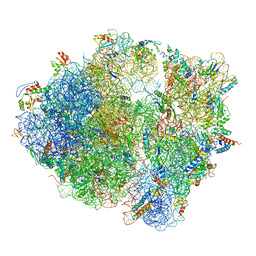 | | Structure of the Bacillus subtilis hibernating 100S ribosome reveals the basis for 70S dimerization. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Beckert, B, Abdelshahid, M, Schaefer, H, Steinchen, W, Arenz, S, Berninghausen, O, Beckmann, R, Bange, G, Turgay, K, Wilson, D.N. | | Deposit date: | 2017-03-29 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Bacillus subtilis hibernating 100S ribosome reveals the basis for 70S dimerization.
EMBO J., 36, 2017
|
|
5NLA
 
 | |
9HVJ
 
 | |
7OG5
 
 | |
5AFF
 
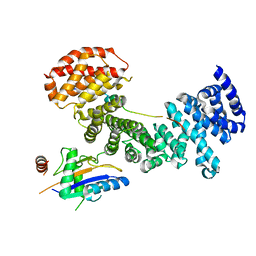 | | Symportin 1 chaperones 5S RNP assembly during ribosome biogenesis by occupying an essential rRNA binding site | | Descriptor: | RIBOSOMAL PROTEIN L11, RIBOSOMAL PROTEIN L5, SYMPORTIN 1 | | Authors: | Calvino, F.R, Kharde, S, Wild, K, Bange, G, Sinning, I. | | Deposit date: | 2015-01-21 | | Release date: | 2015-04-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.398 Å) | | Cite: | Symportin 1 Chaperones 5S Rnp Assembly During Ribosome Biogenesis by Occupying an Essential Rrna-Binding Site.
Nat.Commun., 6, 2015
|
|
9G79
 
 | |
9EN1
 
 | |
7M7K
 
 | |
7O9F
 
 | | Bacillus subtilis Ffh in complex with ppGpp | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, MAGNESIUM ION, Signal recognition particle protein | | Authors: | Czech, L, Mais, C.-N, Bange, G. | | Deposit date: | 2021-04-16 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Inhibition of SRP-dependent protein secretion by the bacterial alarmone (p)ppGpp.
Nat Commun, 13, 2022
|
|
7O9I
 
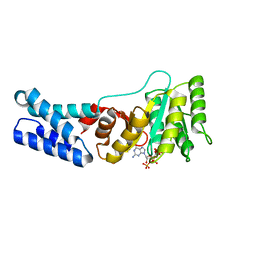 | | Escherichia coli Ffh in complex with pppGpp | | Descriptor: | Signal recognition particle protein, guanosine 5'-(tetrahydrogen triphosphate) 3'-(trihydrogen diphosphate) | | Authors: | Czech, L, Mais, C.-N, Bange, G. | | Deposit date: | 2021-04-16 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Inhibition of SRP-dependent protein secretion by the bacterial alarmone (p)ppGpp.
Nat Commun, 13, 2022
|
|
7O9G
 
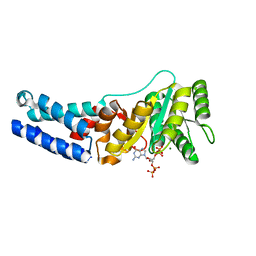 | | Escherichia coli Ffh in complex with ppGpp | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, MAGNESIUM ION, Signal recognition particle protein | | Authors: | Czech, L, Mais, C.-N, Bange, G. | | Deposit date: | 2021-04-16 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Inhibition of SRP-dependent protein secretion by the bacterial alarmone (p)ppGpp.
Nat Commun, 13, 2022
|
|
7O9H
 
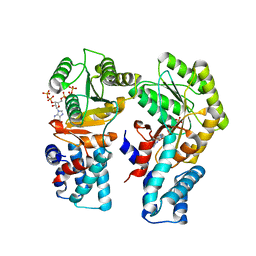 | | Escherichia coli FtsY in complex with pppGpp | | Descriptor: | Signal recognition particle receptor FtsY, guanosine 5'-(tetrahydrogen triphosphate) 3'-(trihydrogen diphosphate) | | Authors: | Czech, L, Mais, C.-N, Bange, G. | | Deposit date: | 2021-04-16 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of SRP-dependent protein secretion by the bacterial alarmone (p)ppGpp.
Nat Commun, 13, 2022
|
|
7O5B
 
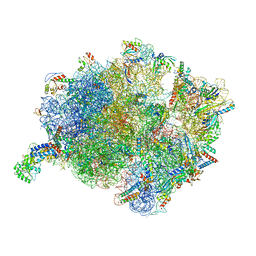 | | Cryo-EM structure of a Bacillus subtilis MifM-stalled ribosome-nascent chain complex with (p)ppGpp-SRP bound | | Descriptor: | 16S rRNA (1533-MER), 23S rRNA (2887-MER), 30S ribosomal protein S10, ... | | Authors: | Kratzat, H, Czech, L, Berninghausen, O, Bange, G, Beckmann, R. | | Deposit date: | 2021-04-08 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Inhibition of SRP-dependent protein secretion by the bacterial alarmone (p)ppGpp.
Nat Commun, 13, 2022
|
|
7P1W
 
 | |
5JRL
 
 | |
5JQF
 
 | | Crystal structure of the lasso peptide Sphingopyxin I (SpI) | | Descriptor: | Sphingopyxin I | | Authors: | Fage, C.D, Hegemann, J.D, Harms, K, Bange, G, Marahiel, M.A. | | Deposit date: | 2016-05-04 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Structure and Mechanism of the Sphingopyxin I Lasso Peptide Isopeptidase.
Angew. Chem. Int. Ed. Engl., 55, 2016
|
|
5JAK
 
 | | Crystal structure of the flagellar assembly factor FliW | | Descriptor: | Flagellar assembly factor FliW | | Authors: | Altegoer, F, Bange, G. | | Deposit date: | 2016-04-12 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis for the CsrA-dependent modulation of translation initiation by an ancient regulatory protein.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5J8Q
 
 | |
5DED
 
 | | Crystal structure of the small alarmone synthethase 1 from Bacillus subtilis bound to its product pppGpp | | Descriptor: | GTP pyrophosphokinase YjbM, MAGNESIUM ION, guanosine 5'-(tetrahydrogen triphosphate) 3'-(trihydrogen diphosphate) | | Authors: | Steinchen, W, Schuhmacher, J.S, Altegoer, F, Bange, G. | | Deposit date: | 2015-08-25 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.942 Å) | | Cite: | Catalytic mechanism and allosteric regulation of an oligomeric (p)ppGpp synthetase by an alarmone.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5JRK
 
 | | Crystal Structure of the Sphingopyxin I Lasso Peptide Isopeptidase SpI-IsoP (SeMet-derived) | | Descriptor: | Dipeptidyl aminopeptidases/acylaminoacyl-peptidases-like protein, beta-D-glucopyranose | | Authors: | Fage, C.D, Hegemann, J.D, Bange, G, Marahiel, M.A. | | Deposit date: | 2016-05-06 | | Release date: | 2016-09-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and Mechanism of the Sphingopyxin I Lasso Peptide Isopeptidase.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5DEC
 
 | |
5DMD
 
 | | Crystal structure of the flagellar assembly factor FliW | | Descriptor: | Flagellar assembly factor FliW | | Authors: | Altegoer, F, Bange, G. | | Deposit date: | 2015-09-08 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis for the CsrA-dependent modulation of translation initiation by an ancient regulatory protein.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5DMB
 
 | |
9H4E
 
 | |
9H3I
 
 | | trans-aconitate decarboxylase Tad1- wild type | | Descriptor: | Trans-aconitate decarboxylase 1 | | Authors: | Zheng, L, Bang, G. | | Deposit date: | 2024-10-16 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Mechanistic and structural insights into the itaconate-producing trans -aconitate decarboxylase Tad1.
Pnas Nexus, 4, 2025
|
|
