4CI6
 
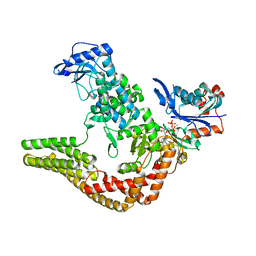 | | Mechanisms of crippling actin-dependent phagocytosis by YopO | | Descriptor: | ACTIN, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Lee, W.L, Grimes, J.M, Robinson, R.C. | | Deposit date: | 2013-12-06 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Yersinia Effector Yopo Uses Actin as Bait to Phosphorylate Proteins that Regulate Actin Polymerization.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4H9V
 
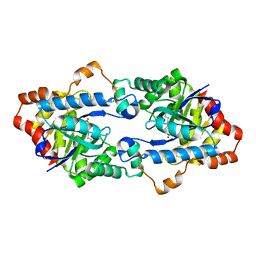 | | Structure of Geobacillus kaustophilus lactonase, mutant E101G/R230C with Zn2+ | | Descriptor: | FE (III) ION, HYDROXIDE ION, Phosphotriesterase, ... | | Authors: | Xue, B, Chow, J.Y, Yew, W.S, Robinson, R.C. | | Deposit date: | 2012-09-25 | | Release date: | 2012-11-07 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (1.971 Å) | | Cite: | Structural evidence of a productive active site architecture for an evolved quorum-quenching GKL lactonase.
Biochemistry, 52, 2013
|
|
4H9T
 
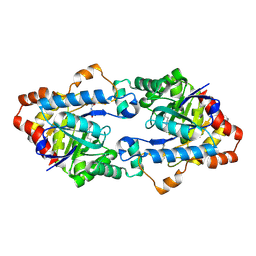 | | Structure of Geobacillus kaustophilus lactonase, mutant E101N with bound N-butyryl-DL-homoserine lactone | | Descriptor: | FE (III) ION, MANGANESE (II) ION, N-[(3S)-2-oxotetrahydrofuran-3-yl]butanamide, ... | | Authors: | Xue, B, Chow, J.Y, Yew, W.S, Robinson, R.C. | | Deposit date: | 2012-09-24 | | Release date: | 2012-11-07 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Structural evidence of a productive active site architecture for an evolved quorum-quenching GKL lactonase.
Biochemistry, 52, 2013
|
|
4H9U
 
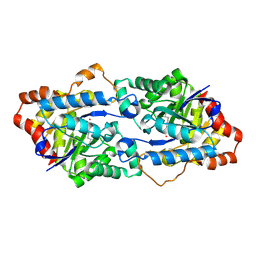 | | Structure of Geobacillus kaustophilus lactonase, wild-type with Zn2+ | | Descriptor: | FE (III) ION, HYDROXIDE ION, Phosphotriesterase, ... | | Authors: | Xue, B, Chow, J.Y, Yew, W.S, Robinson, R.C. | | Deposit date: | 2012-09-25 | | Release date: | 2012-11-07 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural evidence of a productive active site architecture for an evolved quorum-quenching GKL lactonase.
Biochemistry, 52, 2013
|
|
4HA0
 
 | | Structure of Geobacillus kaustophilus lactonase, mutant R230D with Zn2+ | | Descriptor: | FE (III) ION, HYDROXIDE ION, Phosphotriesterase, ... | | Authors: | Xue, B, Chow, J.Y, Yew, W.S, Robinson, R.C. | | Deposit date: | 2012-09-25 | | Release date: | 2012-11-07 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural evidence of a productive active site architecture for an evolved quorum-quenching GKL lactonase.
Biochemistry, 52, 2013
|
|
4H9X
 
 | | Structure of Geobacillus kaustophilus lactonase, mutant E101G/R230C/D266N with Zn2+ and bound N-butyryl-DL-homoserine lactone | | Descriptor: | FE (III) ION, HYDROXIDE ION, N-[(3S)-2-oxotetrahydrofuran-3-yl]butanamide, ... | | Authors: | Xue, B, Chow, J.Y, Yew, W.S, Robinson, R.C. | | Deposit date: | 2012-09-25 | | Release date: | 2012-11-07 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural evidence of a productive active site architecture for an evolved quorum-quenching GKL lactonase.
Biochemistry, 52, 2013
|
|
4H9Y
 
 | | Structure of Geobacillus kaustophilus lactonase, mutant E101N with Zn2+ | | Descriptor: | FE (III) ION, HYDROXIDE ION, Phosphotriesterase, ... | | Authors: | Xue, B, Chow, J.Y, Yew, W.S, Robinson, R.C. | | Deposit date: | 2012-09-25 | | Release date: | 2012-11-07 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.085 Å) | | Cite: | Structural evidence of a productive active site architecture for an evolved quorum-quenching GKL lactonase.
Biochemistry, 52, 2013
|
|
4H9Z
 
 | | Structure of Geobacillus kaustophilus lactonase, mutant E101N with Mn2+ | | Descriptor: | FE (III) ION, MANGANESE (II) ION, Phosphotriesterase | | Authors: | Xue, B, Chow, J.Y, Yew, W.S, Robinson, R.C. | | Deposit date: | 2012-09-25 | | Release date: | 2012-11-07 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural evidence of a productive active site architecture for an evolved quorum-quenching GKL lactonase.
Biochemistry, 52, 2013
|
|
2GV0
 
 | |
4YJY
 
 | |
4YH2
 
 | |
5A1M
 
 | | Crystal structure of calcium-bound human adseverin domain A3 | | Descriptor: | ADSEVERIN, CALCIUM ION | | Authors: | Chumnarnsilpa, S, Robinson, R.C, Grimes, J.M, Leyrat, C. | | Deposit date: | 2015-05-01 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Calcium-Controlled Conformational Choreography in the N-Terminal Half of Adseverin.
Nat.Commun., 6, 2015
|
|
5A1K
 
 | |
5B6I
 
 | | Structure of fluorinase from Streptomyces sp. MA37 | | Descriptor: | ADENOSINE, Fluorinase, METHIONINE | | Authors: | Xue, B, Robinson, R.C. | | Deposit date: | 2016-05-29 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Directed Evolution of a Fluorinase for Improved Fluorination Efficiency with a Non-native Substrate
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5CE3
 
 | | The Yersinia YopO - actin complex with MgADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Lee, W.L, Singaravelu, P, Grimes, J.M, Robinson, R.C. | | Deposit date: | 2015-07-06 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | The Yersinia YopO - actin complex with MgADP
To Be Published
|
|
3AHT
 
 | | Crystal structure of rice BGlu1 E176Q mutant in complex with laminaribiose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, SULFATE ION, ... | | Authors: | Chuenchor, W, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Svasti, J, Ketudat Cairns, J.R. | | Deposit date: | 2010-04-29 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
3AHV
 
 | | Semi-active E176Q mutant of rice bglu1 covalent complex with 2-deoxy-2-fluoroglucoside | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-deoxy-2-fluoro-alpha-D-glucopyranose, Beta-glucosidase 7, ... | | Authors: | Chuenchor, W, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Svasti, J, Ketudat Cairns, J.R. | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
3B9E
 
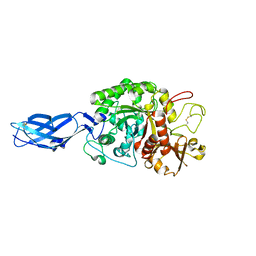 | | Crystal structure of inactive mutant E315M chitinase A from Vibrio harveyi | | Descriptor: | Chitinase A | | Authors: | Songsiriritthigul, C, Pantoom, S, Aguda, A.H, Robinson, R.C, Suginta, W. | | Deposit date: | 2007-11-05 | | Release date: | 2008-04-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of Vibrio harveyi chitinase A complexed with chitooligosaccharides: implications for the catalytic mechanism
J.Struct.Biol., 162, 2008
|
|
3B8S
 
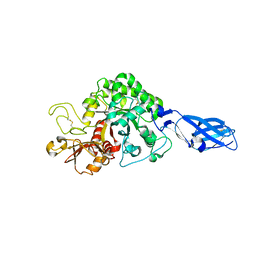 | | Crystal structure of wild-type chitinase A from Vibrio harveyi | | Descriptor: | Chitinase A | | Authors: | Songsiriritthigul, C, Pantoom, S, Aguda, A.H, Robinson, R.C, Suginta, W. | | Deposit date: | 2007-11-01 | | Release date: | 2008-04-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of Vibrio harveyi chitinase A complexed with chitooligosaccharides: implications for the catalytic mechanism
J.Struct.Biol., 162, 2008
|
|
3B9A
 
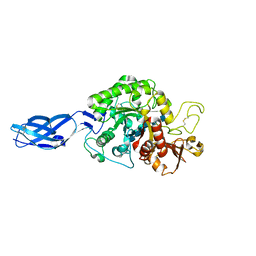 | | Crystal structure of Vibrio harveyi chitinase A complexed with hexasaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase A | | Authors: | Songsiriritthigul, C, Pantoom, S, Aguda, A.H, Robinson, R.C, Suginta, W. | | Deposit date: | 2007-11-03 | | Release date: | 2008-04-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of Vibrio harveyi chitinase A complexed with chitooligosaccharides: implications for the catalytic mechanism
J.Struct.Biol., 162, 2008
|
|
3OJG
 
 | | Structure of an inactive lactonase from Geobacillus kaustophilus with bound N-butyryl-DL-homoserine lactone | | Descriptor: | FE (III) ION, N-[(3S)-2-oxotetrahydrofuran-3-yl]butanamide, Phosphotriesterase, ... | | Authors: | Xue, B, Chow, J.Y, Tung, A, Robinson, R.C. | | Deposit date: | 2010-08-22 | | Release date: | 2010-10-27 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Directed evolution of a thermostable quorum-quenching lactonase from the amidohydrolase superfamily
J.Biol.Chem., 285, 2010
|
|
2BVE
 
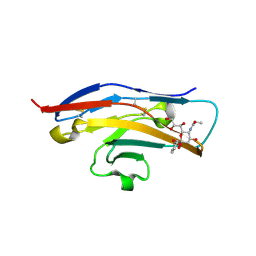 | | Structure of the N-terminal of Sialoadhesin in complex with 2-Phenyl- Prop5Ac | | Descriptor: | SIALOADHESIN, benzyl 3,5-dideoxy-5-(propanoylamino)-D-glycero-alpha-D-galacto-non-2-ulopyranosidonic acid | | Authors: | Zaccai, N.R, May, A.P, Robinson, R.C, Burtnick, L.D, Crocker, P, Brossmer, R, Kelm, S, Jones, E.Y. | | Deposit date: | 2005-06-27 | | Release date: | 2006-07-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic and in Silico Analysis of the Sialoside-Binding Characteristics of the Siglec Sialoadhesin.
J.Mol.Biol., 365, 2007
|
|
2FF3
 
 | | Crystal structure of Gelsolin domain 1:N-wasp V2 motif hybrid in complex with actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Xue, B, Aguda, A.H, Robinson, R.C. | | Deposit date: | 2005-12-19 | | Release date: | 2006-03-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structural Basis of Actin Interaction with Multiple WH2/beta-Thymosin Motif-Containing Proteins
Structure, 14, 2006
|
|
2FF6
 
 | | Crystal structure of Gelsolin domain 1:ciboulot domain 2 hybrid in complex with actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Aguda, A.H, Xue, B, Robinson, R.C. | | Deposit date: | 2005-12-19 | | Release date: | 2006-03-21 | | Last modified: | 2018-05-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Structural Basis of Actin Interaction with Multiple WH2/beta-Thymosin Motif-Containing Proteins
Structure, 14, 2006
|
|
2FH3
 
 | | C-terminal half of gelsolin soaked in low calcium at pH 8 | | Descriptor: | CALCIUM ION, Gelsolin | | Authors: | Chumnarnsilpa, S, Loonchanta, A, Xue, B, Choe, H, Urosev, D, Wang, H, Burtnick, L.D, Robinson, R.C. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Calcium ion exchange in crystalline gelsolin
J.Mol.Biol., 357, 2006
|
|
