1Q4X
 
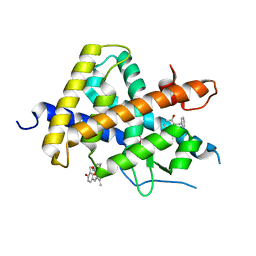 | | Crystal Structure of Human Thyroid Hormone Receptor beta LBD in complex with specific agonist GC-24 | | Descriptor: | Thyroid hormone receptor beta-1, [4-(3-BENZYL-4-HYDROXYBENZYL)-3,5-DIMETHYLPHENOXY]ACETIC ACID | | Authors: | Borngraeber, S, Budny, M.J, Chiellini, G, Cunha-Lima, S.T, Togashi, M, Webb, P, Baxter, J.D, Scanlan, T.S, Fletterick, R.J. | | Deposit date: | 2003-08-04 | | Release date: | 2004-02-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ligand selectivity by seeking hydrophobicity in thyroid hormone receptor.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1EAP
 
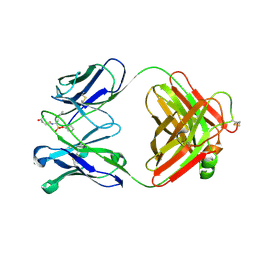 | | CRYSTAL STRUCTURE OF A CATALYTIC ANTIBODY WITH A SERINE PROTEASE ACTIVE SITE | | Descriptor: | IGG2B-KAPPA 17E8 FAB (HEAVY CHAIN), IGG2B-KAPPA 17E8 FAB (LIGHT CHAIN), PHENYL[1-(N-SUCCINYLAMINO)PENTYL]PHOSPHONATE | | Authors: | Zhou, G.W, Guo, J, Huang, W, Scanlan, T.S, Fletterick, R.J. | | Deposit date: | 1994-08-10 | | Release date: | 1994-12-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a catalytic antibody with a serine protease active site.
Science, 265, 1994
|
|
1HGX
 
 | |
1ID5
 
 | |
1I6I
 
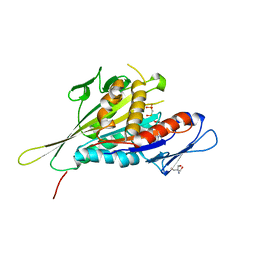 | | CRYSTAL STRUCTURE OF THE KIF1A MOTOR DOMAIN COMPLEXED WITH MG-AMPPCP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, KINESIN-LIKE PROTEIN KIF1A, MAGNESIUM ION, ... | | Authors: | Kikkawa, M, Sablin, E.P, Okada, Y, Yajima, H, Fletterick, R.J, Hirokawa, N. | | Deposit date: | 2001-03-02 | | Release date: | 2001-05-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Switch-based mechanism of kinesin motors
Nature, 411, 2001
|
|
1I5S
 
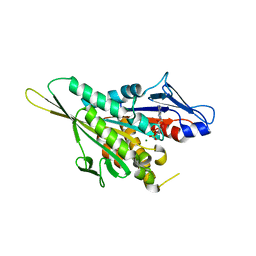 | | CRYSTAL STRUCTURE OF THE KIF1A MOTOR DOMAIN COMPLEXED WITH MG-ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KINESIN-LIKE PROTEIN KIF1A, MAGNESIUM ION | | Authors: | Kikkawa, M, Sablin, E.P, Okada, Y, Yajima, H, Fletterick, R.J, Hirokawa, N. | | Deposit date: | 2001-02-28 | | Release date: | 2001-05-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Switch-based mechanism of kinesin motors
Nature, 411, 2001
|
|
1IA0
 
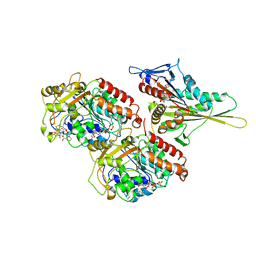 | | KIF1A HEAD-MICROTUBULE COMPLEX STRUCTURE IN ATP-FORM | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, KINESIN-LIKE PROTEIN KIF1A, ... | | Authors: | Kikkawa, M, Sablin, E.P, Okada, Y, Yajima, H, Fletterick, R.J, Hirokawa, N. | | Deposit date: | 2001-03-22 | | Release date: | 2002-03-22 | | Last modified: | 2021-10-27 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Switch-based Mechanism of Kinesin Motors
Nature, 411, 2001
|
|
1EZU
 
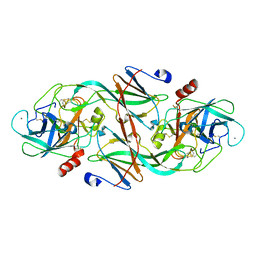 | | ECOTIN Y69F, D70P BOUND TO D102N TRYPSIN | | Descriptor: | CALCIUM ION, ECOTIN, TRYPSIN II, ... | | Authors: | Gillmor, S.A, Takeuchi, T, Yang, S.Q, Craik, C.S, Fletterick, R.J. | | Deposit date: | 2000-05-11 | | Release date: | 2000-06-23 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Compromise and accommodation in ecotin, a dimeric macromolecular inhibitor of serine proteases.
J.Mol.Biol., 299, 2000
|
|
1EZS
 
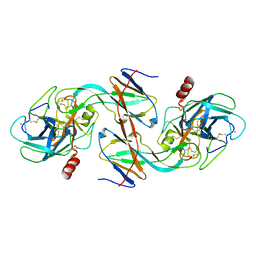 | | CRYSTAL STRUCTURE OF ECOTIN MUTANT M84R, W67A, G68A, Y69A, D70A BOUND TO RAT ANIONIC TRYPSIN II | | Descriptor: | CALCIUM ION, ECOTIN, TRYPSIN II, ... | | Authors: | Gillmor, S.A, Takeuchi, T, Yang, S.Q, Craik, C.S, Fletterick, R.J. | | Deposit date: | 2000-05-11 | | Release date: | 2000-06-23 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Compromise and accommodation in ecotin, a dimeric macromolecular inhibitor of serine proteases.
J.Mol.Biol., 299, 2000
|
|
1IFG
 
 | | CRYSTAL STRUCTURE OF A MONOMERIC FORM OF GENERAL PROTEASE INHIBITOR, ECOTIN IN ABSENCE OF A PROTEASE | | Descriptor: | ECOTIN | | Authors: | Eggers, C.T, Wang, S.X, Fletterick, R.J, Craik, C.S. | | Deposit date: | 2001-04-12 | | Release date: | 2001-05-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The role of ecotin dimerization in protease inhibition.
J.Mol.Biol., 308, 2001
|
|
2YHX
 
 | |
3BKI
 
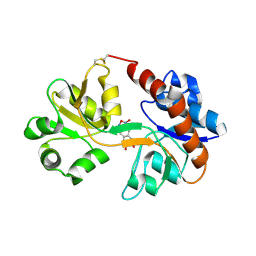 | | Crystal Structure of the GluR2 ligand binding core (S1S2J) in complex with FQX at 1.87 Angstroms | | Descriptor: | Glutamate receptor 2, [1,2,5]oxadiazolo[3,4-g]quinoxaline-6,7(5H,8H)-dione 1-oxide | | Authors: | Cruz, L, Estebanez-Perpina, E, Pfaff, S, Borngraeber, S, Bao, N, Fletterick, R, England, P. | | Deposit date: | 2007-12-06 | | Release date: | 2008-09-16 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | 6-Azido-7-nitro-1,4-dihydroquinoxaline-2,3-dione (ANQX) forms an irreversible bond to the active site of the GluR2 AMPA receptor.
J.Med.Chem., 51, 2008
|
|
3D57
 
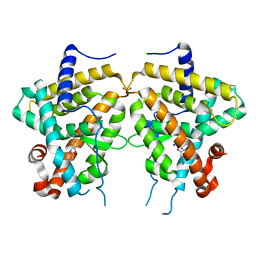 | | TR Variant D355R | | Descriptor: | SULFATE ION, Thyroid hormone receptor beta, [4-(4-HYDROXY-3-IODO-PHENOXY)-3,5-DIIODO-PHENYL]-ACETIC ACID | | Authors: | Jouravel, N. | | Deposit date: | 2008-05-15 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis for dimer formation of TRbeta variant D355R.
Proteins, 75, 2008
|
|
2TRM
 
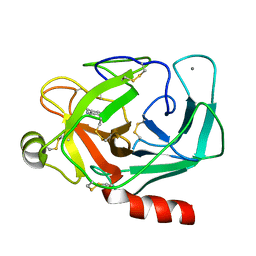 | |
7GPB
 
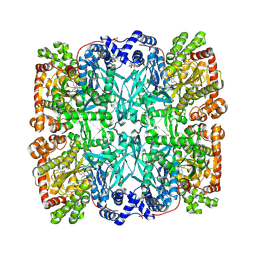 | | STRUCTURAL MECHANISM FOR GLYCOGEN PHOSPHORYLASE CONTROL BY PHOSPHORYLATION AND AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCOGEN PHOSPHORYLASE B, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Barford, D, Hu, S.-H, Johnson, L.N. | | Deposit date: | 1990-11-13 | | Release date: | 1992-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural mechanism for glycogen phosphorylase control by phosphorylation and AMP.
J.Mol.Biol., 218, 1991
|
|
3TX7
 
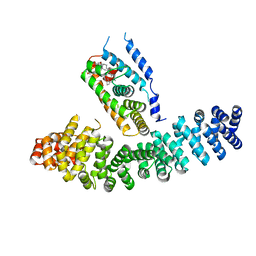 | | Crystal structure of LRH-1/beta-catenin complex | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, Catenin beta-1, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Yumoto, F, Fletterick, R. | | Deposit date: | 2011-09-22 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural basis of coactivation of liver receptor homolog-1 by beta-catenin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
9GPB
 
 | |
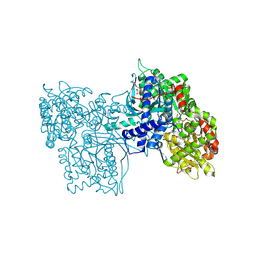
6GPB
 
 | |
1AIM
 
 | |
1PYG
 
 | |
1LOX
 
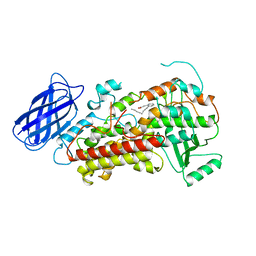 | | RABBIT RETICULOCYTE 15-LIPOXYGENASE | | Descriptor: | (2E)-3-(2-OCT-1-YN-1-YLPHENYL)ACRYLIC ACID, 15-LIPOXYGENASE, FE (II) ION | | Authors: | Gillmor, S.A, Villasenor, A, Fletterick, R.J, Sigal, E, Browner, M.F. | | Deposit date: | 1997-10-06 | | Release date: | 1998-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of mammalian 15-lipoxygenase reveals similarity to the lipases and the determinants of substrate specificity.
Nat.Struct.Biol., 4, 1997
|
|
4OY2
 
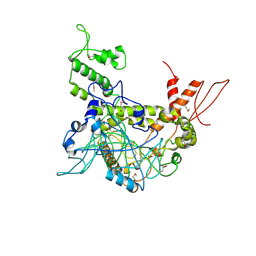 | | Crystal structure of TAF1-TAF7, a TFIID subcomplex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Transcription initiation factor TFIID subunit 1, Transcription initiation factor TFIID subunit 7, ... | | Authors: | Bhattacharya, S, Lou, X, Rajashankar, K, Jacobson, R.H, Webb, P. | | Deposit date: | 2014-02-10 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional insight into TAF1-TAF7, a subcomplex of transcription factor II D.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2PIR
 
 | | Androgen receptor LBD with small molecule | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, SALICYLALDEHYDE, ... | | Authors: | Estebanez-Perpina, E, Arnold, A.A, Baxter, J.D, Webb, P, Guy, R.K, Fletterick, K.R. | | Deposit date: | 2007-04-13 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A surface on the androgen receptor that allosterically regulates coactivator binding.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2PIW
 
 | | Androgen receptor with small molecule | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor | | Authors: | Estebanez-Perpina, E, Arnold, A.A, Baxter, J.D, Webb, P, Guy, R.K, Fletterick, K.R. | | Deposit date: | 2007-04-13 | | Release date: | 2007-09-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | A surface on the androgen receptor that allosterically regulates coactivator binding.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2PIQ
 
 | | androgen receptor LBD with small molecule | | Descriptor: | 3-[(4-AMINO-1-TERT-BUTYL-1H-PYRAZOLO[3,4-D]PYRIMIDIN-3-YL)METHYL]PHENOL, 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, ... | | Authors: | Estebanez-Perpina, E, Arnold, A.A, Baxter, J.D, Webb, P, Guy, R.K, Fletterick, K.R. | | Deposit date: | 2007-04-13 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A surface on the androgen receptor that allosterically regulates coactivator binding.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
