8UTU
 
 | | KIF1A[1-393] P305L mutant AMP-PNP bound one and two heads bound states merged, in complex with a microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF1A, ... | | Authors: | Benoit, M.P.M.H, Rao, L, Asenjo, A.B, Gennerich, A, Sosa, H. | | Deposit date: | 2023-10-31 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM unveils kinesin KIF1A's processivity mechanism and the impact of its pathogenic variant P305L.
Nat Commun, 15, 2024
|
|
8UTQ
 
 | | KIF1A[1-393] AMP-PNP bound one-head-bound state in complex with a microtubule - class T1L02* | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF1A, ... | | Authors: | Benoit, M.P.M.H, Rao, L, Asenjo, A.B, Gennerich, A, Sosa, H. | | Deposit date: | 2023-10-31 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM unveils kinesin KIF1A's processivity mechanism and the impact of its pathogenic variant P305L.
Nat Commun, 15, 2024
|
|
8UTT
 
 | | KIF1A[1-393] P305L mutant AMP-PNP bound two-heads-bound state in complex with a microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF1A, ... | | Authors: | Benoit, M.P.M.H, Rao, L, Asenjo, A.B, Gennerich, A, Sosa, H. | | Deposit date: | 2023-10-31 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM unveils kinesin KIF1A's processivity mechanism and the impact of its pathogenic variant P305L.
Nat Commun, 15, 2024
|
|
8UTO
 
 | | KIF1A[1-393] AMP-PNP bound two-heads-bound state in complex with a microtubule - class T2L1 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF1A, ... | | Authors: | Benoit, M.P.M.H, Rao, L, Asenjo, A.B, Gennerich, A, Sosa, H. | | Deposit date: | 2023-10-31 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM unveils kinesin KIF1A's processivity mechanism and the impact of its pathogenic variant P305L.
Nat Commun, 15, 2024
|
|
8UTR
 
 | | KIF1A[1-393] ADP bound in complex with a microtubule | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Benoit, M.P.M.H, Rao, L, Asenjo, A.B, Gennerich, A, Sosa, H. | | Deposit date: | 2023-10-31 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM unveils kinesin KIF1A's processivity mechanism and the impact of its pathogenic variant P305L.
Nat Commun, 15, 2024
|
|
8U44
 
 | | CryoEM structure of A/Solomon Islands/3/2006 H1 HA in complex with 05.GC.w2.3C10-H1_SI06 | | Descriptor: | 05.GC.w2.3C10-H1_SI06 Heavy chain, 05.GC.w2.3C10-H1_SI06 Light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Moore, N, Han, J, Ward, A.B, Wilson, I.A. | | Deposit date: | 2023-09-08 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Persistence of germinal center B cell responses after influenza virus vaccination in humans
To Be Published
|
|
6AHX
 
 | | Copper-Sensing Operon Regulator Protein (CsoRGz) | | Descriptor: | Putative cytosolic protein | | Authors: | Normi, M.Y, Mangavelu, A, Sayangku, A.A, Jonet, M.A, Adam, T.C.L, Ali, M.S.M, Rahman, R.N.Z.R.A, Salleh, A.B. | | Deposit date: | 2018-08-21 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallization, Structural Determination and Analysis of Copper-sensing Operon Regulator Protein (CsoRGz) of Geobacillus zalihae Strain T1
To Be Published
|
|
6AWD
 
 | | Structure of 30S (S1 depleted) ribosomal subunit and RNA polymerase complex | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demo, G, Rasouly, A, Vasilyev, N, Loveland, A.B, Diaz-Avalos, R, Grigorieff, N, Nudler, E, Korostelev, A.A. | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Structure of RNA polymerase bound to ribosomal 30S subunit.
Elife, 6, 2017
|
|
6B0C
 
 | | KLP10A-AMPPNP in complex with curved tubulin and a microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein Klp10A, ... | | Authors: | Benoit, M.P.M.H, Asenjo, A.B, Sosa, H. | | Deposit date: | 2017-09-14 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Cryo-EM reveals the structural basis of microtubule depolymerization by kinesin-13s.
Nat Commun, 9, 2018
|
|
6APQ
 
 | | Anti-Marburgvirus Nucleoprotein Single Domain Antibody B | | Descriptor: | Anti-Marburgvirus Nucleoprotein Single Domain Antibody B, CHLORIDE ION, SODIUM ION | | Authors: | Taylor, A.B, Garza, J.A. | | Deposit date: | 2017-08-17 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unveiling a Drift Resistant Cryptotope withinMarburgvirusNucleoprotein Recognized by Llama Single-Domain Antibodies.
Front Immunol, 8, 2017
|
|
6B0L
 
 | | KLP10A-AMPPNP in complex with a microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein Klp10A, ... | | Authors: | Benoit, M.P.M.H, Asenjo, A.B, Sosa, H. | | Deposit date: | 2017-09-14 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Cryo-EM reveals the structural basis of microtubule depolymerization by kinesin-13s.
Nat Commun, 9, 2018
|
|
6B0I
 
 | | Apo KLP10A in complex with a microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein Klp10A, ... | | Authors: | Benoit, M.P.M.H, Asenjo, A.B, Sosa, H. | | Deposit date: | 2017-09-14 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Cryo-EM reveals the structural basis of microtubule depolymerization by kinesin-13s.
Nat Commun, 9, 2018
|
|
6B51
 
 | | Schistosoma haematobium (Blood Fluke) Sulfotransferase, Y54F Mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase | | Authors: | Taylor, A.B. | | Deposit date: | 2017-09-27 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Why does oxamniquine kill Schistosoma mansoni and not S. haematobium and S. japonicum?
Int.J.Parasitol., 2020
|
|
6BDP
 
 | | Schistosoma mansoni (Blood Fluke) Sulfotransferase/CIDD-0000071 (Compound 9c) Complex | | Descriptor: | (4-{[(3R)-1-benzylpyrrolidin-3-yl]amino}-2-nitrophenyl)methanol, ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase oxamniquine resistance protein | | Authors: | Taylor, A.B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Design, Synthesis, and Characterization of Novel Small Molecules as Broad Range Antischistosomal Agents.
ACS Med Chem Lett, 9, 2018
|
|
6AWC
 
 | | Structure of 30S ribosomal subunit and RNA polymerase complex in rotated state | | Descriptor: | 16S rRNA, 30S ribosomal protein S1, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Rasouly, A, Vasilyev, N, Loveland, A.B, Diaz-Avalos, R, Grigorieff, N, Nudler, E, Korostelev, A.A. | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structure of RNA polymerase bound to ribosomal 30S subunit.
Elife, 6, 2017
|
|
6B1V
 
 | | Crystal structure of Ps i-CgsB C78S in complex with i-neocarratetraose | | Descriptor: | 1,2-ETHANEDIOL, 3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose-(1-4)-3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose, CALCIUM ION, ... | | Authors: | Hettle, A.G, Boraston, A.B. | | Deposit date: | 2017-09-19 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | The Molecular Basis of Polysaccharide Sulfatase Activity and a Nomenclature for Catalytic Subsites in this Class of Enzyme.
Structure, 26, 2018
|
|
6B4X
 
 | | Schistosoma mansoni (Blood Fluke) Sulfotransferase, F39Y Mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-3'-5'-DIPHOSPHATE, SODIUM ION, ... | | Authors: | Taylor, A.B. | | Deposit date: | 2017-09-27 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Why does oxamniquine kill Schistosoma mansoni and not S. haematobium and S. japonicum?
Int.J.Parasitol., 2020
|
|
6BDC
 
 | |
6BDR
 
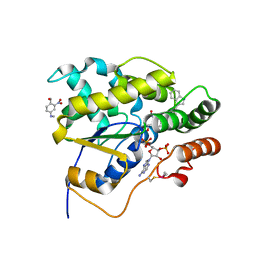 | | Schistosoma mansoni (Blood Fluke) Sulfotransferase/CIDD-0000206 (Compound 9f) Complex | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase oxamniquine resistance protein, [4-({(3R)-1-[(1H-indol-3-yl)methyl]pyrrolidin-3-yl}amino)-2-nitrophenyl]methanol | | Authors: | Taylor, A.B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Design, Synthesis, and Characterization of Novel Small Molecules as Broad Range Antischistosomal Agents.
ACS Med Chem Lett, 9, 2018
|
|
6BDS
 
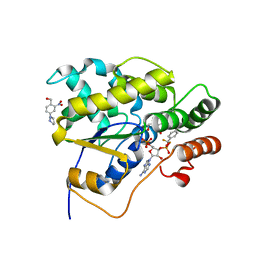 | | Schistosoma mansoni (Blood Fluke) Sulfotransferase/CIDD-0000204 (Compound 11f) Complex | | Descriptor: | (2-nitro-4-{[(3S)-1-{[4-(trifluoromethyl)phenyl]methyl}pyrrolidin-3-yl]amino}phenyl)methanol, ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase oxamniquine resistance protein | | Authors: | Taylor, A.B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Design, Synthesis, and Characterization of Novel Small Molecules as Broad Range Antischistosomal Agents.
ACS Med Chem Lett, 9, 2018
|
|
6B53
 
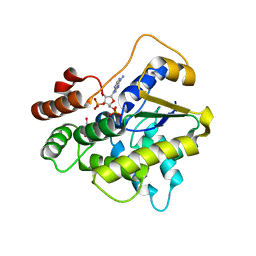 | | Schistosoma haematobium (Blood Fluke) Sulfotransferase, S166T Mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase | | Authors: | Taylor, A.B. | | Deposit date: | 2017-09-27 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Why does oxamniquine kill Schistosoma mansoni and not S. haematobium and S. japonicum?
Int.J.Parasitol., 2020
|
|
6APP
 
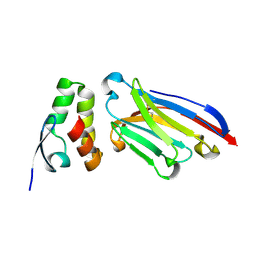 | |
6B0K
 
 | | Crystal structure of Ps i-CgsB C78S in complex with k-carrapentaose | | Descriptor: | 1,2-ETHANEDIOL, 3,6-anhydro-D-galactose, 4-O-sulfo-beta-D-galactopyranose, ... | | Authors: | Hettle, A, Boraston, A.B. | | Deposit date: | 2017-09-14 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Molecular Basis of Polysaccharide Sulfatase Activity and a Nomenclature for Catalytic Subsites in this Class of Enzyme.
Structure, 26, 2018
|
|
6B0J
 
 | | Crystal structure of Ps i-CgsB in complex with k-i-k-neocarrahexaose | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose, ... | | Authors: | Hettle, A.G, Boraston, A.B. | | Deposit date: | 2017-09-14 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Molecular Basis of Polysaccharide Sulfatase Activity and a Nomenclature for Catalytic Subsites in this Class of Enzyme.
Structure, 26, 2018
|
|
6B4Y
 
 | | Schistosoma mansoni (Blood Fluke) Sulfotransferase/Oxamniquine Complex, F39Y Mutant | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase oxamniquine resistance protein, {(2S)-7-nitro-2-[(propan-2-ylamino)methyl]-1,2,3,4-tetrahydroquinolin-6-yl}methanol | | Authors: | Taylor, A.B. | | Deposit date: | 2017-09-27 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Why does oxamniquine kill Schistosoma mansoni and not S. haematobium and S. japonicum?
Int.J.Parasitol., 2020
|
|
