3HIC
 
 | |
3HDO
 
 | |
3HJG
 
 | | Crystal structure of putative alpha-ribazole-5'-phosphate phosphatase CobC FROM Vibrio parahaemolyticus | | Descriptor: | Putative alpha-ribazole-5'-phosphate phosphatase CobC, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Morano, C, Rutter, M, Iizuka, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-21 | | Release date: | 2009-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of putative alpha-ribazole-5' -phosphate phosphatase FROM Vibrio parahaemolyticus
To be Published
|
|
3HD5
 
 | |
3HEB
 
 | |
3G2E
 
 | | Structure of putative OORC subunit of 2-oxoglutarate:acceptor oxidoreductase from Campylobacter jejuni | | Descriptor: | GLYCEROL, OORC subunit of 2-oxoglutarate:acceptor oxidoreductase | | Authors: | Ramagopal, U.A, Toro, R, Miller, S, Gilmore, M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-01-31 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of putative OORC subunit of 2-oxoglutarate:acceptor oxidoreductase from
Campylobacter jejuni
To be Published
|
|
3GD6
 
 | | Crystal structure of divergent enolase from Oceanobacillus iheyensis complexed with phosphate | | Descriptor: | Muconate cycloisomerase, PHOSPHATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-23 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of divergent enolase from Oceanobacillus iheyensis complexed with phosphate.
To be Published
|
|
3GDO
 
 | |
3GFO
 
 | | Structure of cbiO1 from clostridium perfringens: Part of the ABC transporter complex cbiONQ. | | Descriptor: | Cobalt import ATP-binding protein cbiO 1, SULFATE ION | | Authors: | Ramagopal, U.A, Morano, C, Toro, R, Dickey, M, Do, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of cbiO1 from clostridium perfringens: Part of the ABC transporter complex cbiONQ
To be published
|
|
3CU5
 
 | | Crystal structure of a two component transcriptional regulator AraC from Clostridium phytofermentans ISDg | | Descriptor: | Two component transcriptional regulator, AraC family | | Authors: | Malashkevich, V.N, Toro, R, Wasserman, S.R, Meyer, A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-15 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a two component transcriptional regulator AraC from Clostridium phytofermentans ISDg.
To be Published
|
|
3CG0
 
 | | Crystal structure of signal receiver domain of modulated diguanylate cyclase from Desulfovibrio desulfuricans G20, an example of alternate folding | | Descriptor: | Response regulator receiver modulated diguanylate cyclase with PAS/PAC sensor | | Authors: | Patskovsky, Y, Bonanno, J.B, Romero, R, Gilmore, M, Chang, S, Groshong, C, Koss, J, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-04 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Signal Receiver Domain of Modulated Diguanylate Cyclase from Desulfovibrio desulfuricans.
To be Published
|
|
3GFG
 
 | | Structure of putative oxidoreductase yvaA from Bacillus subtilis in triclinic form | | Descriptor: | Uncharacterized oxidoreductase yvaA | | Authors: | Ramagopal, U.A, Toro, R, Gilmore, M, Chang, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-26 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of putative oxidoreductase yvaA from Bacillus subtilis in triclinic form.
To be published
|
|
3G12
 
 | | Crystal structure of a putative lactoylglutathione lyase from Bdellovibrio bacteriovorus | | Descriptor: | Putative lactoylglutathione lyase, SULFATE ION | | Authors: | Patskovsky, Y, Madegowda, M, Gilmore, M, Chang, S, Maletic, M, Smith, D, Sauder, J.M, Burley, S.K, Swaminathan, S, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-01-29 | | Release date: | 2009-02-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of a putative lactoylglutathione lyase from Bdellovibrio bacteriovorus
To be Published
|
|
3L60
 
 | | Crystal structure of branched-chain alpha-keto acid dehydrogenase subunit e2 from mycobacterium tuberculosis | | Descriptor: | BRANCHED-CHAIN ALPHA-KETO ACID DEHYDROGENASE, UNKNOWN LIGAND | | Authors: | Zencheck, W.D, Bonanno, J.B, Patskovsky, Y, Toro, R, Freeman, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-12-22 | | Release date: | 2010-01-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Branched-Chain Alpha-Keto Acid Dehydrogenase Subunit E2 from Mycobacterium Tuberculosis
To be Published
|
|
3I4F
 
 | | Structure of putative 3-oxoacyl-reductase from bacillus thuringiensis | | Descriptor: | 3-oxoacyl-[acyl-carrier protein] reductase | | Authors: | Ramagopal, U.A, Kim, J, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-01 | | Release date: | 2009-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of putative 3-oxoacyl-reductase from bacillus thuringiensis
To be published
|
|
3LUA
 
 | |
3FND
 
 | |
3FI9
 
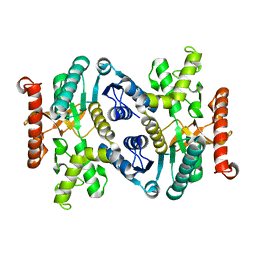 | | Crystal structure of malate dehydrogenase from Porphyromonas gingivalis | | Descriptor: | Malate dehydrogenase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Miller, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-12-11 | | Release date: | 2008-12-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of malate dehydrogenase from Porphyromonas gingivalis
To be Published
|
|
3I5T
 
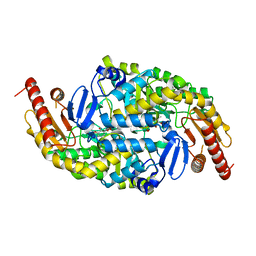 | | CRYSTAL STRUCTURE OF AMINOTRANSFERASE PRK07036 FROM Rhodobacter sphaeroides KD131 | | Descriptor: | Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Do, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-06 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF AMINOTRANSFERASE PRK07036 FROM Rhodobacter sphaeroides
To be Published
|
|
3CTA
 
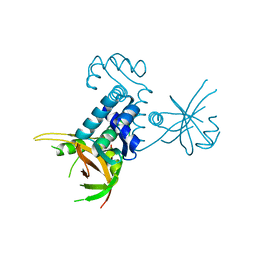 | | Crystal structure of riboflavin kinase from Thermoplasma acidophilum | | Descriptor: | Riboflavin kinase | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Mendoza, M, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-11 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of riboflavin kinase from Thermoplasma acidophilum.
To be Published
|
|
3CTP
 
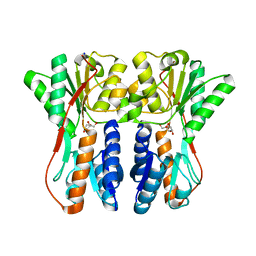 | | Crystal structure of periplasmic binding protein/LacI transcriptional regulator from Alkaliphilus metalliredigens QYMF complexed with D-xylulofuranose | | Descriptor: | Periplasmic binding protein/LacI transcriptional regulator, SODIUM ION, beta-D-xylulofuranose | | Authors: | Malashkevich, V.N, Toro, R, Wasserman, S.R, Meyer, A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-14 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal structure of periplasmic binding protein/LacI transcriptional regulator from Alkaliphilus metalliredigens QYMF complexed with L-xylulose.
To be Published
|
|
3MSY
 
 | |
3M3P
 
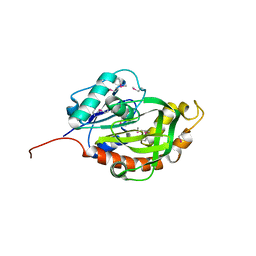 | | Crystal structure of glutamine amido transferase from Methylobacillus Flagellatus | | Descriptor: | Glutamine amido transferase | | Authors: | Fedorov, A.A, Domagalski, M, Fedorov, E.V, Burley, S.K, Minor, W, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-09 | | Release date: | 2010-03-23 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of glutamine amido transferase from Methylobacillus Flagellatus
To be Published
|
|
3G1W
 
 | |
3G13
 
 | |
