8PRA
 
 | |
2V92
 
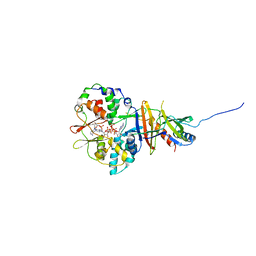 | | Crystal structure of the regulatory fragment of mammalian AMPK in complexes with ATP-AMP | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Heath, R, Saiu, P, Leiper, F.C, Leone, P, Jing, C, Walker, P.A, Haire, L, Eccleston, J.F, Davis, C.T, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2007-08-20 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for AMP Binding to Mammalian AMP-Activated Protein Kinase
Nature, 449, 2007
|
|
8PR9
 
 | |
8E68
 
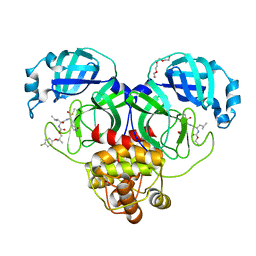 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a p-fluorodimethyl oxybenzene inhibitor | | Descriptor: | (1S,2S)-2-[(N-{[2-(4-fluorophenoxy)-2-methylpropoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, N~2~-{[2-(4-fluorophenoxy)-2-methylpropoxy]carbonyl}-N-{(1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-1-sulfanylpropan-2-yl}-L-leucinamide, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Miller, M.J, Groutas, W.C. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
8E63
 
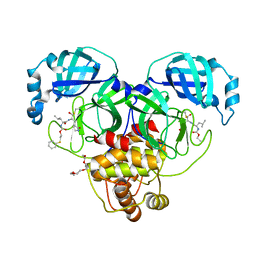 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a phenyl sulfane inhibitor | | Descriptor: | (1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-[(N-{[2-(phenylsulfanyl)ethoxy]carbonyl}-L-leucyl)amino]propane-1-sulfonic acid, 2-phenylsulfanylethyl ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Madden, T.K, Groutas, W.C. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
8PRQ
 
 | |
8PRR
 
 | |
8PRH
 
 | |
2VCO
 
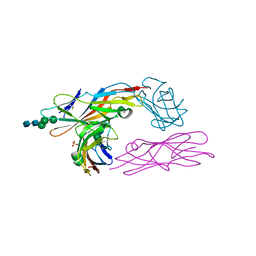 | | Crystal structure of the fimbrial adhesin FimH in complex with its high-mannose epitope | | Descriptor: | NICKEL (II) ION, PROTEIN FIMH, SULFATE ION, ... | | Authors: | Wellens, A, Garofalo, C, Nguyen, H, Wyns, L, De Greve, H, Hultgren, S.J, Bouckaert, J. | | Deposit date: | 2007-09-26 | | Release date: | 2008-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Intervening with Urinary Tract Infections Using Anti-Adhesives Based on the Crystal Structure of the Fimh-Oligomannose-3 Complex.
Plos One, 3, 2008
|
|
8PRS
 
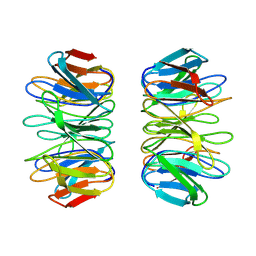 | |
4E80
 
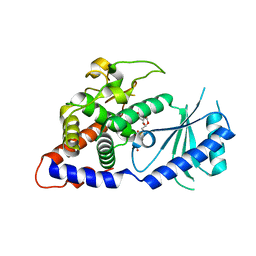 | | Structural Basis for the Activity of a Cytoplasmic RNA Terminal U-transferase | | Descriptor: | Poly(A) RNA polymerase protein cid1, URIDINE 5'-TRIPHOSPHATE | | Authors: | Yates, L.A, Fleurdepine, S, Rissland, O.S, DeColibus, L, Harlos, K, Norbury, C.J, Gilbert, R.J.C. | | Deposit date: | 2012-03-19 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural basis for the activity of a cytoplasmic RNA terminal uridylyl transferase.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1K3D
 
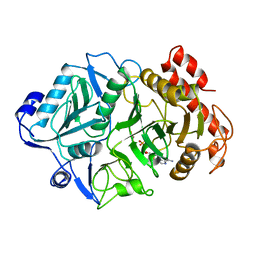 | | Phosphoenolpyruvate carboxykinase in complex with ADP and AlF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Sudom, A.M, Prasad, L, Goldie, H, Delbaere, L.T.J. | | Deposit date: | 2001-10-02 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The phosphoryl-transfer mechanism of Escherichia coli phosphoenolpyruvate carboxykinase from the use of AlF(3).
J.Mol.Biol., 314, 2001
|
|
8PU2
 
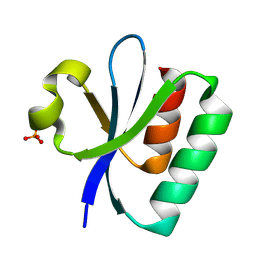 | |
8PU4
 
 | | FaRel2 bound to the ATP analogue, APCPP | | Descriptor: | MAGNESIUM ION, RelA/SpoT domain-containing protein, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-[[oxidanyl(phosphonooxy)phosphoryl]methyl]phosphinic acid | | Authors: | Garcia-Pino, A, Talavera Perez, A, Dominguez Molina, L. | | Deposit date: | 2023-07-16 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | FaRel2 bound to the ATP analogue, APCPP
To Be Published
|
|
8PU3
 
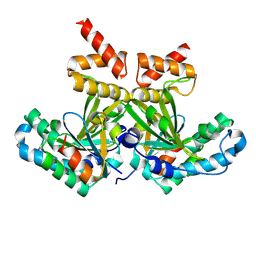 | |
7NKY
 
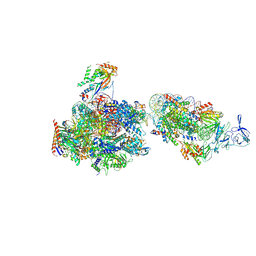 | | RNA Polymerase II-Spt4/5-nucleosome-FACT structure | | Descriptor: | Chromatin elongation factor SPT4, DNA (138-MER), DNA (148-MER), ... | | Authors: | Farnung, L, Ochmann, M, Engeholm, M, Cramer, P. | | Deposit date: | 2021-02-19 | | Release date: | 2021-07-07 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of nucleosome transcription mediated by Chd1 and FACT.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7NN3
 
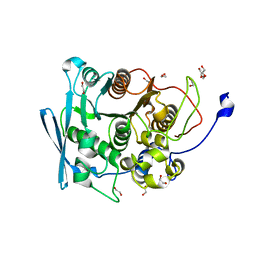 | | A carbohydrate esterase family 15 (CE15) glucuronoyl esterase from Caldicellulosiruptor kristjansonii | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Beta-xylanase, ... | | Authors: | Krska, D, Mazurkewich, S, Navarro Poulsen, J, Larsbrink, J, Lo Leggio, L. | | Deposit date: | 2021-02-24 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88948345 Å) | | Cite: | Structural and Functional Analysis of a Multimodular Hyperthermostable Xylanase-Glucuronoyl Esterase from Caldicellulosiruptor kristjansonii .
Biochemistry, 60, 2021
|
|
8PRI
 
 | |
8PRL
 
 | |
2V22
 
 | | REPLACE: A strategy for Iterative Design of Cyclin Binding Groove Inhibitors | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2, N~2~-{[1-(4-CHLOROPHENYL)-5-METHYL-1H-1,2,4-TRIAZOL-3-YL]CARBONYL}-N~5~-(DIAMINOMETHYLIDENE)-L-ORNITHYL-L-LEUCYL-L-ISOLEUCYL-4-FLUORO-L-PHENYLALANINAMIDE | | Authors: | Andrews, M.J, Kontopidis, G, McInnes, C, Plater, A, Innes, L, Cowan, A, Jewsbury, P, Fischer, P.M. | | Deposit date: | 2007-05-31 | | Release date: | 2008-01-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Replace: A Strategy for Iterative Design of Cyclin- Binding Groove Inhibitors
Chembiochem, 7, 2006
|
|
6YU2
 
 | | Crystal structure of MhsT in complex with L-isoleucine | | Descriptor: | ISOLEUCINE, SODIUM ION, Sodium-dependent transporter, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
1K3G
 
 | | NMR Solution Structure of Oxidized Cytochrome c-553 from Bacillus pasteurii | | Descriptor: | HEME C, cytochrome c-553 | | Authors: | Banci, L, Bertini, I, Ciurli, S, Dikiy, A, Dittmer, J, Rosato, A, Sciara, G, Thompsett, A.R. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure, backbone mobility, and homology modeling of c-type cytochromes from gram-positive bacteria.
Chembiochem, 3, 2002
|
|
6YU3
 
 | | Crystal structure of MhsT in complex with L-phenylalanine | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, GLYCEROL, PHENYLALANINE, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
8Q3W
 
 | | ATP-bound IstB in complex to duplex DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (48-MER) Target DNA FW, DNA (48-MER) Traget DNA Rv, ... | | Authors: | de la Gandara, A, Spinola-Amilibia, M, Araujo-Bazan, L, Nunez-Ramirez, R, Berger, J.M, Arias-Palomo, E. | | Deposit date: | 2023-08-04 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Molecular basis for transposase activation by a dedicated AAA+ ATPase.
Nature, 630, 2024
|
|
8PRO
 
 | |
