1V8S
 
 | | Crystal structure analusis of the ADP-ribose pyrophosphatase complexed with AMP and Mg | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADP-ribose pyrophosphatase, MAGNESIUM ION | | Authors: | Yoshiba, S, Ooga, T, Nakagawa, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-14 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal
J.Biol.Chem., 279, 2004
|
|
1VBM
 
 | | Crystal structure of the Escherichia coli tyrosyl-tRNA synthetase complexed with Tyr-AMS | | Descriptor: | 5'-O-[N-(L-TYROSYL)SULFAMOYL]ADENOSINE, SULFATE ION, Tyrosyl-tRNA synthetase | | Authors: | Kobayashi, T, Sakamoto, K, Takimura, T, Kamata, K, Sekine, R, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-27 | | Release date: | 2005-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural snapshots of the KMSKS loop rearrangement for amino acid activation by bacterial tyrosyl-tRNA synthetase.
J.Mol.Biol., 346, 2005
|
|
1V8I
 
 | | Crystal Structure Analysis of the ADP-ribose pyrophosphatase | | Descriptor: | ADP-ribose pyrophosphatase | | Authors: | Yoshiba, S, Ooga, T, Nakagawa, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-09 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal
J.Biol.Chem., 279, 2004
|
|
1V5S
 
 | | Solution structure of kinase associated domain 1 of mouse MAP/microtubule affinity-regulating kinase 3 | | Descriptor: | MAP/microtubule affinity-regulating kinase 3 | | Authors: | Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-25 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of kinase associated domain 1 of mouse MAP/microtubule affinity-regulating kinase 3
To be Published
|
|
1V8W
 
 | | Crystal structure analysis of the ADP-ribose pyrophosphatase of E82Q mutant, complexed with SO4 and Zn | | Descriptor: | ADP-ribose pyrophosphatase, SULFATE ION, ZINC ION | | Authors: | Yoshiba, S, Ooga, T, Nakagawa, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-15 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal
J.Biol.Chem., 279, 2004
|
|
1V96
 
 | |
1VBK
 
 | | Crystal structure of PH1313 from Pyrococcus horikoshii Ot3 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, hypothetical protein PH1313 | | Authors: | Sugahara, M, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-27 | | Release date: | 2005-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Purification, crystallization and preliminary crystallographic analysis of the putative thiamine-biosynthesis protein PH1313 from Pyrococcus horikoshii OT3
Acta Crystallogr.,Sect.F, 63, 2007
|
|
1V5H
 
 | | Crystal Structure of Human Cytoglobin (Ferric Form) | | Descriptor: | Cytoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugimoto, H, Makino, M, Sawai, H, Kawada, N, Yoshizato, K, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-23 | | Release date: | 2004-06-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of human cytoglobin for ligand binding.
J.Mol.Biol., 339, 2004
|
|
1V5R
 
 | | Solution Structure of the Gas2 Domain of the Growth Arrest Specific 2 Protein | | Descriptor: | Growth-arrest-specific protein 2, ZINC ION | | Authors: | Miyamoto, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-25 | | Release date: | 2005-01-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Gas2 Domain of the Growth Arrest Specific 2 Protein
To be Published
|
|
1V6H
 
 | |
1V32
 
 | | Solution structure of the SWIB/MDM2 domain of the hypothetical protein At5g08430 from Arabidopsis thaliana | | Descriptor: | hypothetical protein RAFL09-47-K03 | | Authors: | Yoneyama, M, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-24 | | Release date: | 2004-04-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SWIB/MDM2 domain of the hypothetical protein At5g08430 from Arabidopsis thaliana
To be Published
|
|
1VB7
 
 | |
1WEN
 
 | | Solution structure of PHD domain in ING1-like protein BAC25079 | | Descriptor: | ZINC ION, inhibitor of growth family, member 4; ING1-like protein | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-25 | | Release date: | 2004-11-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of PHD domain in ING1-like protein BAC25079
To be Published
|
|
1WLO
 
 | |
1WR2
 
 | |
1WTY
 
 | |
1WXB
 
 | | Solution structure of the SH3 domain from human epidermal growth factor receptor pathway substrate 8-like protein | | Descriptor: | Epidermal growth factor receptor pathway substrate 8-like protein | | Authors: | Chikayama, E, Kigawa, T, Muto, Y, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-20 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain from human epidermal growth factor receptor pathway substrate 8-like protein
To be Published
|
|
1X43
 
 | |
1X4J
 
 | | Solution structure of RING finger in RING finger protein 38 | | Descriptor: | RING finger protein 38, ZINC ION | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RING finger in RING finger protein 38
To be Published
|
|
1WEK
 
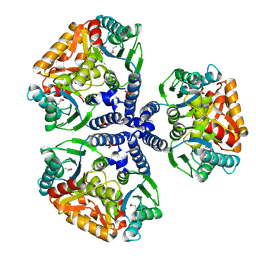 | | Crystal structure of the conserved hypothetical protein TT1465 from Thermus thermophilus HB8 | | Descriptor: | PHOSPHATE ION, hypothetical protein TT1465 | | Authors: | Kukimoto-Niino, M, Murayama, K, Kato-Murayama, M, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-25 | | Release date: | 2004-11-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of possible lysine decarboxylases from Thermus thermophilus HB8
Protein Sci., 13, 2004
|
|
1WGP
 
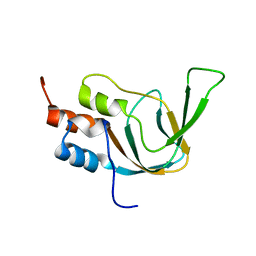 | | Solution structure of the cNMP-binding domain from Arabidopsis thaliana cyclic nucleotide-regulated ion channel | | Descriptor: | Probable cyclic nucleotide-gated ion channel 6 | | Authors: | Chikayama, E, Nameki, N, Kigawa, T, Koshiba, S, Inoue, M, Tomizawa, T, Kobayashi, N, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cNMP-binding domain from Arabidopsis thaliana cyclic nucleotide-regulated ion channel
To be Published
|
|
1WI7
 
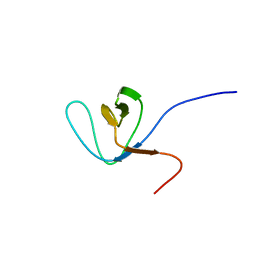 | |
1WNL
 
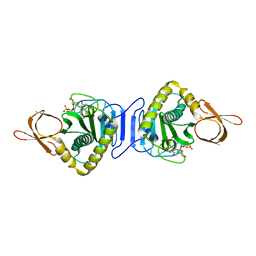 | |
1WXQ
 
 | |
1X1M
 
 | | Solution Structure of the N-terminal Ubiquitin-like Domain in Mouse Ubiquitin-like Protein SB132 | | Descriptor: | Ubiquitin-like protein SB132 | | Authors: | Zhao, C, Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-04-07 | | Release date: | 2005-10-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the N-terminal Ubiquitin-like Domain in Mouse Ubiquitin-like Protein SB132
To be Published
|
|
