5FE9
 
 | | Crystal structure of human PCAF bromodomain in complex with compound SL1122 (compound 13) | | Descriptor: | 1,2-ETHANEDIOL, Histone acetyltransferase KAT2B, ~{N}-(1,4-dimethyl-2-oxidanylidene-quinolin-7-yl)methanesulfonamide | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
5FE6
 
 | | Crystal structure of human PCAF bromodomain in complex with fragment ZB1916 (fragment 10) | | Descriptor: | (4-azanylpiperidin-1-yl)-cyclopropyl-methanone, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
5FE0
 
 | | Crystal structure of human PCAF bromodomain in complex with acetyllysine | | Descriptor: | Histone acetyltransferase KAT2B, N(6)-ACETYLLYSINE | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
5FE7
 
 | | Crystal structure of human PCAF bromodomain in complex with fragment ZB2216 (fragment 11) | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-hydroxyethyl)-3-methyl-6,7-dihydro-5~{H}-indazol-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure-Based Identification of Inhibitory Fragments Targeting the p300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 59, 2016
|
|
3EJC
 
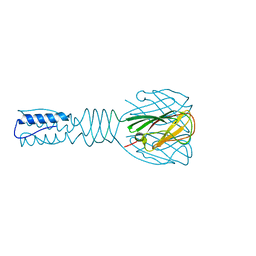 | | Full length Receptor Binding Protein from Lactococcal phage TP901-1 | | Descriptor: | Baseplate protein (BPP) | | Authors: | Spinelli, S, Lichiere, J, Blangy, S, Sciara, G, Cambillau, C, Campanacci, V. | | Deposit date: | 2008-09-18 | | Release date: | 2009-10-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and molecular assignment of lactococcal phage TP901-1 baseplate.
J.Biol.Chem., 285, 2010
|
|
7PUQ
 
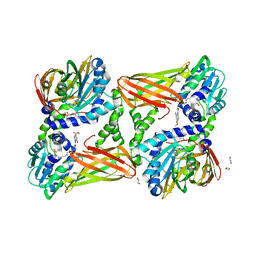 | | CARM1 in complex with EML982 | | Descriptor: | 1,2-ETHANEDIOL, Histone-arginine methyltransferase CARM1, methyl 6-[4-[[~{N}-[3-[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]propyl]carbamimidoyl]amino]butylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-09-30 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
7PPY
 
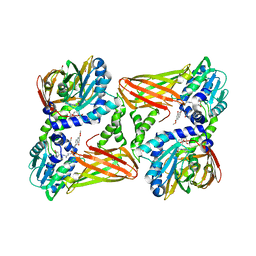 | | CARM1 in complex with EML709 | | Descriptor: | 1,2-ETHANEDIOL, Histone-arginine methyltransferase CARM1, methyl 6-[4-[[~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]carbamimidoyl]amino]butylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | Authors: | Marechal, N, Cura, V, Bonnefond, L, Troffer-Charlier, N, Cavarelli, J. | | Deposit date: | 2021-09-15 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
7PV6
 
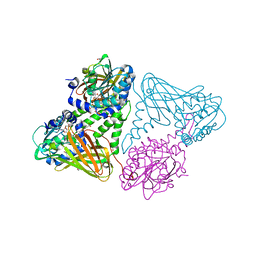 | | CARM1 in complex with EML734 | | Descriptor: | 1,2-ETHANEDIOL, Histone-arginine methyltransferase CARM1, TETRAETHYLENE GLYCOL, ... | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-10-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
7PU8
 
 | | CARM1 in complex with EML980 | | Descriptor: | 1,2-ETHANEDIOL, Histone-arginine methyltransferase CARM1, methyl 6-[4-[[N-[2-[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]ethyl]carbamimidoyl]amino]butylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-09-28 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
7PPQ
 
 | | CARM1 in complex with EML736 | | Descriptor: | 1,2-ETHANEDIOL, Histone-arginine methyltransferase CARM1, methyl 6-[5-[[~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]carbamimidoyl]amino]pentylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | Authors: | Marechal, N, Cura, V, Bonnefond, L, Troffer-Charlier, N, Cavarelli, J. | | Deposit date: | 2021-09-14 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
7PUC
 
 | | CARM1 in complex with EML981 | | Descriptor: | 1,2-ETHANEDIOL, Histone-arginine methyltransferase CARM1, methyl 6-[4-[[~{N}-[(~{E})-3-[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]prop-2-enyl]carbamimidoyl]amino]butylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | Authors: | Marechal, N, Cura, V, Bonnefond, L, Troffer-Charlier, N, Cavarelli, J. | | Deposit date: | 2021-09-29 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
4ZE9
 
 | | Se-PBP AccA from A. tumefaciens C58 in complex with agrocinopine A | | Descriptor: | 2-O-phosphono-alpha-L-arabinopyranose, ABC transporter substrate-binding protein, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | El Sahili, A, Guimaraes, B.G, Morera, S. | | Deposit date: | 2015-04-20 | | Release date: | 2015-08-19 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A Pyranose-2-Phosphate Motif Is Responsible for Both Antibiotic Import and Quorum-Sensing Regulation in Agrobacterium tumefaciens.
Plos Pathog., 11, 2015
|
|
4NYF
 
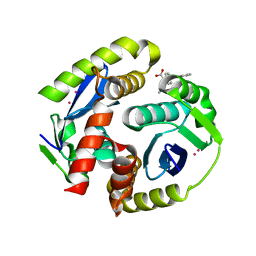 | | HIV integrase in complex with inhibitor | | Descriptor: | (2S)-tert-butoxy[4-(4-chlorophenyl)-2-methylquinolin-3-yl]ethanoic acid, CADMIUM ION, Integrase | | Authors: | Coulombe, R, Fader, L. | | Deposit date: | 2013-12-10 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of BI 224436, a Noncatalytic Site Integrase Inhibitor (NCINI) of HIV-1.
ACS Med Chem Lett, 5, 2014
|
|
4ZED
 
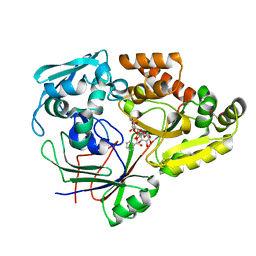 | |
7XGR
 
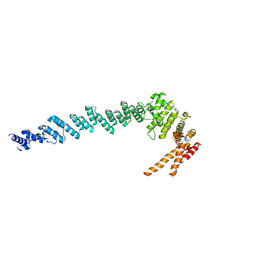 | |
7XDT
 
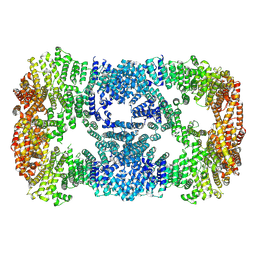 | |
4ZE8
 
 | | PBP AccA from A. tumefaciens C58 | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter, substrate binding protein (Agrocinopines A and B), ... | | Authors: | El Sahili, A, Morera, S. | | Deposit date: | 2015-04-20 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A Pyranose-2-Phosphate Motif Is Responsible for Both Antibiotic Import and Quorum-Sensing Regulation in Agrobacterium tumefaciens.
Plos Pathog., 11, 2015
|
|
4ZEI
 
 | |
1DEF
 
 | |
4ZEC
 
 | | PBP AccA from A. tumefaciens C58 in complex with agrocin 84 | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter, substrate binding protein (Agrocinopines A and B), ... | | Authors: | El Sahili, A, Morera, S. | | Deposit date: | 2015-04-20 | | Release date: | 2015-08-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Pyranose-2-Phosphate Motif Is Responsible for Both Antibiotic Import and Quorum-Sensing Regulation in Agrobacterium tumefaciens.
Plos Pathog., 11, 2015
|
|
4P0I
 
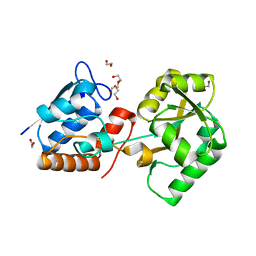 | | Structure of the PBP NocT | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Nopaline-binding periplasmic protein | | Authors: | Vigouroux, A, Morera, S. | | Deposit date: | 2014-02-21 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Agrobacterium uses a unique ligand-binding mode for trapping opines and acquiring a competitive advantage in the niche construction on plant host.
Plos Pathog., 10, 2014
|
|
6FIB
 
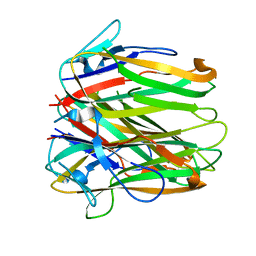 | | Structure of human 4-1BB ligand | | Descriptor: | Tumor necrosis factor ligand superfamily member 9, Tumor necrosis factor ligand superfamily member 9,4-1BBL -CH/CL fusion, Tumor necrosis factor ligand superfamily member 9,Uncharacterized protein | | Authors: | Joseph, C, Claus, C, Ferrara, C, von Hirschheydt, T, Prince, C, Funk, D, Klein, C, Benz, J. | | Deposit date: | 2018-01-17 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Tumor-targeted 4-1BB agonists for combination with T cell bispecific antibodies as off-the-shelf therapy.
Sci Transl Med, 11, 2019
|
|
7RLS
 
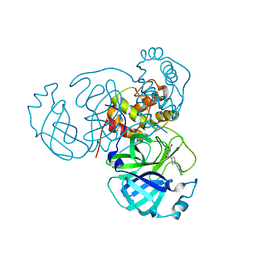 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-68 | | Descriptor: | 3C-like proteinase, 6-[4-(3,4,5-trichlorophenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-26 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RNK
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-71 | | Descriptor: | 3C-like proteinase, 6-{4-[3-chloro-4-(hydroxymethyl)phenyl]piperazine-1-carbonyl}pyrimidine-2,4(3H,5H)-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-29 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RM2
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with Mcule-CSR-494190-S1 | | Descriptor: | 3C-like proteinase, 6-[4-(3,5-dichloro-4-methylphenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-26 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
