3OP4
 
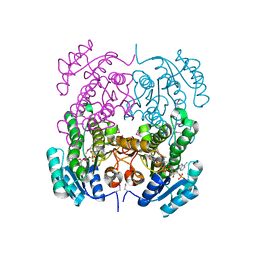 | | Crystal structure of putative 3-ketoacyl-(acyl-carrier-protein) reductase from Vibrio cholerae O1 biovar eltor str. N16961 in complex with NADP+ | | Descriptor: | 3-oxoacyl-[acyl-carrier protein] reductase, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hou, J, Chruszcz, M, Onopriyenko, O, Grimshaw, S, Porebski, P, Zheng, H, Savchenko, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-22 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Dissecting the Structural Elements for the Activation of beta-Ketoacyl-(Acyl Carrier Protein) Reductase from Vibrio cholerae.
J.Bacteriol., 198, 2015
|
|
3RSH
 
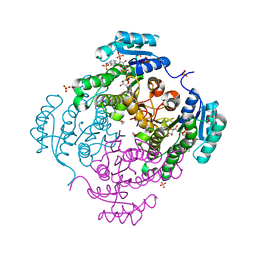 | | Structure of 3-ketoacyl-(acyl-carrier-protein)reductase (FabG) from Vibrio cholerae O1 complexed with NADP+ (space group P62) | | Descriptor: | 1,2-ETHANEDIOL, 3-oxoacyl-[acyl-carrier protein] reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hou, J, Chruszcz, M, Cooper, D.R, Grabowski, M, Zheng, H, Osinski, T, Shumilin, I, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-02 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Dissecting the Structural Elements for the Activation of beta-Ketoacyl-(Acyl Carrier Protein) Reductase from Vibrio cholerae.
J.Bacteriol., 198, 2015
|
|
3RRO
 
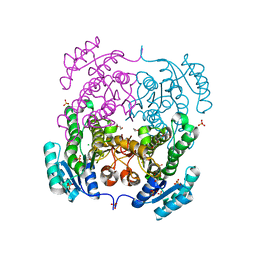 | | Crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase (FabG) from Vibrio cholerae | | Descriptor: | 1,2-ETHANEDIOL, 3-ketoacyl-(acyl-carrier-protein) reductase, CHLORIDE ION, ... | | Authors: | Hou, J, Chruszcz, M, Cooper, D.R, Grabowski, M, Zheng, H, Osinski, T, Shumilin, I, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-29 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dissecting the Structural Elements for the Activation of beta-Ketoacyl-(Acyl Carrier Protein) Reductase from Vibrio cholerae.
J.Bacteriol., 198, 2015
|
|
3IH2
 
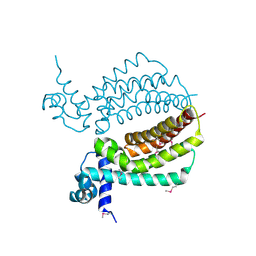 | | TM1030 crystallized at 323K | | Descriptor: | Transcriptional regulator, TetR family | | Authors: | Koclega, K.D, Chruszcz, M, Bujacz, G, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-29 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 'Hot' macromolecular crystals.
Cryst.Growth Des., 10, 2010
|
|
3IH4
 
 | | TM1030 crystallized at 277K | | Descriptor: | Transcriptional regulator, TetR family | | Authors: | Koclega, K.D, Chruszcz, M, Bujacz, G, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-29 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 'Hot' macromolecular crystals.
Cryst.Growth Des., 10, 2010
|
|
3IJW
 
 | | Crystal structure of BA2930 in complex with CoA | | Descriptor: | ACETYL COENZYME *A, Aminoglycoside N3-acetyltransferase, CHLORIDE ION, ... | | Authors: | Klimecka, M.M, Chruszcz, M, Skarina, T, Onopryienko, O, Cymborowski, M, Savchenko, A, Edwards, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-05 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis of a Putative Aminoglycoside N-Acetyltransferase from Bacillus anthracis.
J.Mol.Biol., 410, 2011
|
|
3IH3
 
 | | TM1030 crystallized at 310K | | Descriptor: | Transcriptional regulator, TetR family | | Authors: | Koclega, K.D, Chruszcz, M, Bujacz, G, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-29 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 'Hot' macromolecular crystals.
Cryst.Growth Des., 10, 2010
|
|
7KL8
 
 | | Structure of F420 binding protein Rv1558 from Mycobacterium tuberculosis with F420 bound | | Descriptor: | COENZYME F420, COENZYME F420-3, Deazaflavin-dependent nitroreductase, ... | | Authors: | Lee, B.M, Tan, L.L, Jackson, C.J. | | Deposit date: | 2020-10-29 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.469 Å) | | Cite: | Potency boost of a Mycobacterium tuberculosis dihydrofolate reductase inhibitor by multienzyme F 420 H 2 -dependent reduction.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2FDO
 
 | | Crystal Structure of the Conserved Protein of Unknown Function AF2331 from Archaeoglobus fulgidus DSM 4304 Reveals a New Type of Alpha/Beta Fold | | Descriptor: | Hypothetical protein AF2331 | | Authors: | Wang, S, Kirillova, O, Chruszcz, M, Cymborowski, M.T, Skarina, T, Gorodichtchenskaia, E, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-14 | | Release date: | 2006-01-31 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of the AF2331 protein from Archaeoglobus fulgidus DSM 4304 forms an unusual interdigitated dimer with a new type of alpha + beta fold.
Protein Sci., 18, 2009
|
|
2O5U
 
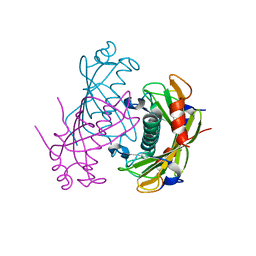 | | Crystal structure of the PA5185 protein from Pseudomonas Aeruginosa strain PAO1- orthorhombic form (C222). | | Descriptor: | Thioesterase | | Authors: | Chruszcz, M, Wang, S, Evdokimova, E, Koclega, K.D, Kudritska, M, Savchenko, A, Edwards, A, Minor, W. | | Deposit date: | 2006-12-06 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Function-biased choice of additives for optimization of protein crystallization - the case of the putative thioesterase PA5185 from Pseudomonas aeruginosa PAO1.
Cryst.Growth Des., 8, 2008
|
|
3V4E
 
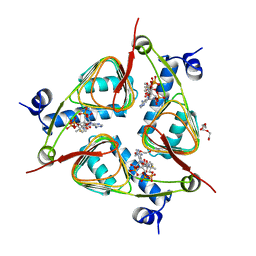 | | Crystal Structure of the galactoside O-acetyltransferase in complex with CoA | | Descriptor: | COENZYME A, DI(HYDROXYETHYL)ETHER, Galactoside O-acetyltransferase, ... | | Authors: | Knapik, A.A, Shumilin, I.A, Luo, H.-B, Chruszcz, M, Zimmerman, M.D, Cymborowski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-14 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biophysical analysis of the putative acetyltransferase SACOL2570 from methicillin-resistant Staphylococcus aureus.
J.Struct.Funct.Genom., 14, 2013
|
|
2O6T
 
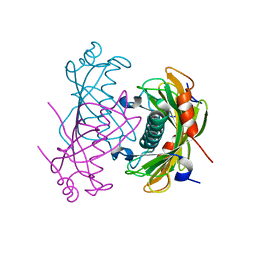 | | Crystal structure of the PA5185 protein from Pseudomonas Aeruginosa strain PAO1- orthorhombic form (P2221). | | Descriptor: | CHLORIDE ION, THIOESTERASE | | Authors: | Chruszcz, M, Koclega, K.D, Evdokimova, E, Cymborowski, M, Kudritska, M, Savchenko, A, Edwards, A, Minor, W. | | Deposit date: | 2006-12-08 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Function-biased choice of additives for optimization of protein crystallization - the case of the putative thioesterase PA5185 from Pseudomonas aeruginosa PAO1.
Cryst.Growth Des., 8, 2008
|
|
2O6U
 
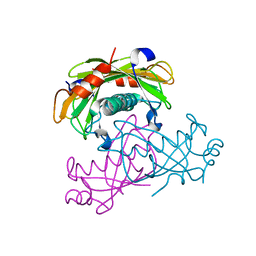 | | Crystal structure of the PA5185 protein from Pseudomonas Aeruginosa strain PAO1- new crystal form. | | Descriptor: | THIOESTERASE | | Authors: | Chruszcz, M, Koclega, K.D, Evdokimova, E, Cymborowski, M, Kudritska, M, Savchenko, A, Edwards, A, Minor, W. | | Deposit date: | 2006-12-08 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Function-biased choice of additives for optimization of protein crystallization - the case of the putative thioesterase PA5185 from Pseudomonas aeruginosa PAO1.
Cryst.Growth Des., 8, 2008
|
|
2O6B
 
 | | Crystal structure of the PA5185 protein from Pseudomonas Aeruginosa strain PAO1- new crystal form. | | Descriptor: | THIOESTERASE | | Authors: | Chruszcz, M, Koclega, K.D, Evdokimova, E, Cymborowski, M, Kudritska, M, Savchenko, A, Edwards, A, Minor, W. | | Deposit date: | 2006-12-07 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Function-biased choice of additives for optimization of protein crystallization - the case of the putative thioesterase PA5185 from Pseudomonas aeruginosa PAO1.
Cryst.Growth Des., 8, 2008
|
|
4KOX
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with Cefalotin | | Descriptor: | 1,2-ETHANEDIOL, CEPHALOTHIN, SULFATE ION, ... | | Authors: | Majorek, K.A, Chruszcz, M, Zimmerman, M.D, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-12 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural, Functional, and Inhibition Studies of a Gcn5-related N-Acetyltransferase (GNAT) Superfamily Protein PA4794: A NEW C-TERMINAL LYSINE PROTEIN ACETYLTRANSFERASE FROM PSEUDOMONAS AERUGINOSA.
J.Biol.Chem., 288, 2013
|
|
3FTT
 
 | | Crystal Structure of the galactoside O-acetyltransferase from Staphylococcus aureus | | Descriptor: | Putative acetyltransferase SACOL2570 | | Authors: | Knapik, A.A, Shumilin, I.A, Cui, H, Xu, X, Chruszcz, M, Zimmerman, M.D, Cymborowski, M, Anderson, W.F, Savchenko, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-01-13 | | Release date: | 2009-03-03 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biophysical analysis of the putative acetyltransferase SACOL2570 from methicillin-resistant Staphylococcus aureus.
J.Struct.Funct.Genom., 14, 2013
|
|
