2PNG
 
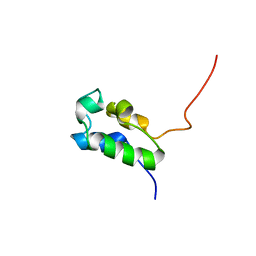 | | Type I rat fatty acid synthase acyl carrier protein (ACP) domain | | Descriptor: | Fatty acid synthase (EC 2.3.1.85) | | Authors: | Ploskon, E.A, Arthur, C.J, Evans, S.E, Williams, C, Crosby, J, Crump, M.P. | | Deposit date: | 2007-04-24 | | Release date: | 2007-06-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A Mammalian type I Fatty Acid synthase acyl carrier protein domain does not sequester acyl chains.
J.Biol.Chem., 283, 2008
|
|
6FXD
 
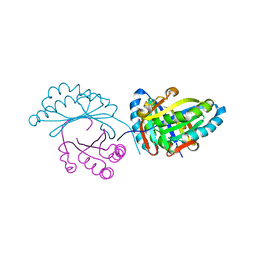 | | Crystal structure of MupZ from Pseudomonas fluorescens | | Descriptor: | MupZ | | Authors: | Wang, L, Parnell, A, Williams, C, Bakar, N.A, van der Kamp, M.W, Simpson, T.J, Race, P.R, Crump, M.P, Willis, C.L. | | Deposit date: | 2018-03-08 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Rieske oxygenase/epoxide hydrolase-catalysed reaction cascade creates oxygen heterocycles in mupirocin biosynthesis
Nat Catal, 2018
|
|
6G1C
 
 | |
6G26
 
 | | The crystal structure of the Burkholderia pseudomallei HicAB complex | | Descriptor: | 1,2-ETHANEDIOL, HicA, HicB, ... | | Authors: | Winter, A.J, Isupov, M.N, Williams, C, Crump, M.P. | | Deposit date: | 2018-03-22 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The molecular basis of protein toxin HicA-dependent binding of the protein antitoxin HicB to DNA.
J. Biol. Chem., 293, 2018
|
|
6G1N
 
 | |
1Q1J
 
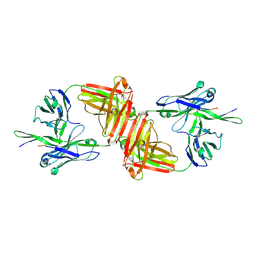 | | Crystal Structure Analysis of anti-HIV-1 Fab 447-52D in complex with V3 peptide | | Descriptor: | Fab 447-52D, heavy chain, light chain, ... | | Authors: | Stanfield, R.L, Gorny, M.K, Williams, C, Zolla-Pazner, S, Wilson, I.A. | | Deposit date: | 2003-07-21 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural rationale for the broad neutralization of HIV-1 by human monoclonal antibody 447-52D.
Structure, 12, 2004
|
|
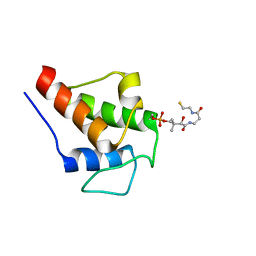
2K0X
 
 | |
2JOL
 
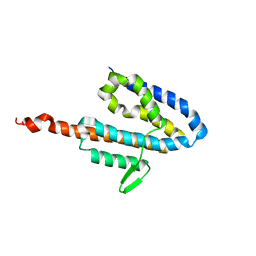 | | Average NMR structure of the catalytic domain of guanine nucleotide exchange factor BopE from Burkholderia pseudomallei | | Descriptor: | Putative G-nucleotide exchange factor | | Authors: | Wu, H, Upadhyay, A, Williams, C, Galyov, E.E, van den Elsen, J.M.H, Bagby, S. | | Deposit date: | 2007-03-14 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The guanine-nucleotide-exchange factor BopE from Burkholderia pseudomallei adopts a compact version of the Salmonella SopE/SopE2 fold and undergoes a closed-to-open conformational change upon interaction with Cdc42
Biochem.J., 411, 2008
|
|
2JOK
 
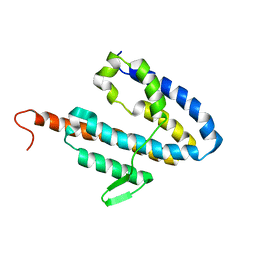 | | NMR structure of the catalytic domain of guanine nucleotide exchange factor BopE from Burkholderia pseudomallei | | Descriptor: | Putative G-nucleotide exchange factor | | Authors: | Wu, H, Upadhyay, A, Williams, C, Galyov, E.E, van den Elsen, J.M.H, Bagby, S. | | Deposit date: | 2007-03-14 | | Release date: | 2007-09-18 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The guanine-nucleotide-exchange factor BopE from Burkholderia pseudomallei adopts a compact version of the Salmonella SopE/SopE2 fold and undergoes a closed-to-open conformational change upon interaction with Cdc42
Biochem.J., 411, 2008
|
|
2KGD
 
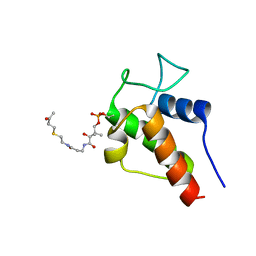 | | NMR Solution Structures of 3-oxo-butyl-ACP, an intermediate mimic from the actinorhodin polyketide synthase in Streptomyces coelicolor | | Descriptor: | Actinorhodin polyketide synthase acyl carrier protein, N~3~-[(2S)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-N-{2-[(3-oxobutyl)sulfanyl]ethyl}-beta-alaninamide | | Authors: | Crump, M.P, Evans, S.E, Williams, C. | | Deposit date: | 2009-03-08 | | Release date: | 2009-04-14 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Probing the Interactions of Early Polyketide Intermediates with the Actinorhodin ACP from S. coelicolor A3(2).
J.Mol.Biol., 389, 2009
|
|
2KGC
 
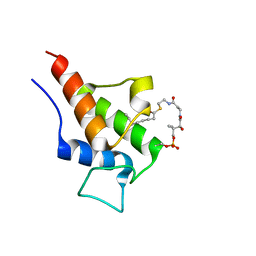 | |
2KG9
 
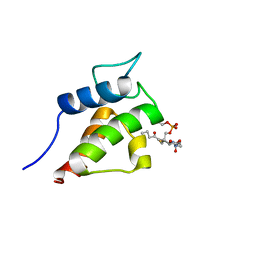 | | NMR Solution Structures of butyryl-ACP (a non-polar, non pathway intermediate) from the actinorhodin polyketide synthase in Streptomyces coelicolor | | Descriptor: | Actinorhodin polyketide synthase acyl carrier protein, THIOBUTYRIC ACID S-{2-[3-(2-HYDROXY-3,3-DIMETHYL-4-PHOSPHONOOXY-BUTYRYLAMINO)-PROPIONYLAMINO]-ETHYL} ESTER | | Authors: | Crump, M.P, Evans, S.E, Williams, C. | | Deposit date: | 2009-03-07 | | Release date: | 2009-04-14 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Probing the Interactions of Early Polyketide Intermediates with the Actinorhodin ACP from S. coelicolor A3(2).
J.Mol.Biol., 389, 2009
|
|
2KGE
 
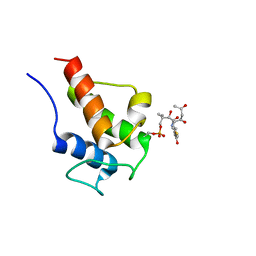 | | NMR Solution Structures of 3,5-dioxohexyl ACP (a triketide mimic) from the actinorhodin polyketide synthase in Streptomyces coelicolor | | Descriptor: | Actinorhodin polyketide synthase acyl carrier protein, N-{2-[(3,5-dioxohexyl)sulfanyl]ethyl}-N~3~-[(2S)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide | | Authors: | Crump, M.P, Evans, S.E, Williams, C. | | Deposit date: | 2009-03-08 | | Release date: | 2009-04-14 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Probing the Interactions of Early Polyketide Intermediates with the Actinorhodin ACP from S. coelicolor A3(2).
J.Mol.Biol., 389, 2009
|
|
2KG8
 
 | | NMR Solution Structures of malonyl ACP from the actinorhodin polyketide synthase in Streptomyces coelicolor | | Descriptor: | 3-{[2-({N-[(2S)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl]sulfanyl}-3-oxopropanoic acid, Actinorhodin polyketide synthase acyl carrier protein | | Authors: | Crump, M.P, Evans, S.E, Williams, C, Eliza, P. | | Deposit date: | 2009-03-06 | | Release date: | 2009-04-14 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Probing the Interactions of Early Polyketide Intermediates with the Actinorhodin ACP from S. coelicolor A3(2).
J.Mol.Biol., 389, 2009
|
|
2KGA
 
 | |
2KR5
 
 | | Solution Structure of an Acyl Carrier Protein Domain from Fungal Type I Polyketide Synthase | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Aflatoxin biosynthesis polyketide synthase | | Authors: | Wattana-amorn, P, Williams, C, Ploskon, E, Cox, R.J, Simpson, T.J, Crosby, J, Crump, M.P. | | Deposit date: | 2009-12-03 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an acyl carrier protein domain from a fungal type I polyketide synthase.
Biochemistry, 49, 2010
|
|
3Q6G
 
 | | Crystal structure of Fab of rhesus mAb 2.5B specific for quaternary neutralizing epitope of HIV-1 gp120 | | Descriptor: | Heavy chain of Fab of rhesus mAb 2.5B, Light chain of Fab of rhesus mAb 2.5B | | Authors: | Spurrier, B, Sampson, J, Totrov, M, Li, H, O'Neal, T, William, C, Robinson, J, Gorny, M.K, Zolla-Pazner, S, Kong, X.P. | | Deposit date: | 2010-12-31 | | Release date: | 2011-05-25 | | Last modified: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural Analysis of Human and Macaque mAbs 2909 and 2.5B: Implications for the Configuration of the Quaternary Neutralizing Epitope of HIV-1 gp120.
Structure, 19, 2011
|
|
3Q6F
 
 | | Crystal structure of Fab of human mAb 2909 specific for quaternary neutralizing epitope of HIV-1 gp120 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Heavy chain of Fab of human mAb 2909, Light chain of Fab of human mAb 2909 | | Authors: | Spurrier, B, Sampson, J, Totrov, M, Li, H, O'Neal, T, William, C, Robinson, J, Gorny, M.K, Zolla-Pazner, S, Kong, X.P. | | Deposit date: | 2010-12-31 | | Release date: | 2011-05-25 | | Last modified: | 2011-10-12 | | Method: | X-RAY DIFFRACTION (3.192 Å) | | Cite: | Structural Analysis of Human and Macaque mAbs 2909 and 2.5B: Implications for the Configuration of the Quaternary Neutralizing Epitope of HIV-1 gp120.
Structure, 19, 2011
|
|
4C26
 
 | | Solution NMR structure of the HicA toxin from Burkholderia pseudomallei | | Descriptor: | HICA | | Authors: | Butt, A, Higman, V.A, Williams, C, Crump, M.P, Hemsley, C, Harmer, N, Titball, R.W. | | Deposit date: | 2013-08-16 | | Release date: | 2014-04-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Hica Toxin from Burkholderia Pseudomallei Has a Role in Persister Cell Formation.
Biochem.J., 459, 2014
|
|
3UJI
 
 | | Crystal structure of anti-HIV-1 V3 Fab 2558 in complex with MN peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Envelope glycoprotein gp160, ... | | Authors: | Kong, X.P. | | Deposit date: | 2011-11-07 | | Release date: | 2011-12-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Human Anti-V3 HIV-1 Monoclonal Antibodies Encoded by the VH5-51/VL Lambda Genes Define a Conserved Antigenic Structure.
Plos One, 6, 2011
|
|
3UJJ
 
 | |
5Y08
 
 | |
7AH0
 
 | | Crystal structure of the de novo designed two-heme binding protein, 4D2 | | Descriptor: | 4D2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hutchins, G.H, Parnell, A.E, Anderson, J.L.R. | | Deposit date: | 2020-09-23 | | Release date: | 2021-10-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | An expandable, modular de novo protein platform for precision redox engineering.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5F14
 
 | | Structure of native hen egg-white lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | McGlone, C, Nix, J.C, Page, R.C. | | Deposit date: | 2015-11-30 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.148 Å) | | Cite: | Investigating the Impact of Polymer Functional Groups on the Stability and Activity of Lysozyme-Polymer Conjugates.
Biomacromolecules, 17, 2016
|
|
5F16
 
 | | CTA-modified hen egg-white lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | McGlone, C, Nix, J.C, Page, R.C. | | Deposit date: | 2015-11-30 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Investigating the Impact of Polymer Functional Groups on the Stability and Activity of Lysozyme-Polymer Conjugates.
Biomacromolecules, 17, 2016
|
|
