5Y0O
 
 | | Crystal structure of apo BsTmcAL | | Descriptor: | UPF0348 protein B4417_3650 | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2017-07-18 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Acetate-dependent tRNA acetylation required for decoding fidelity in protein synthesis.
Nat. Chem. Biol., 14, 2018
|
|
5Y0T
 
 | |
6IW6
 
 | | Crystal structure of the Lin28-interacting module of human TUT4 | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Terminal uridylyltransferase 4,Terminal uridylyltransferase 4, ... | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2018-12-04 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Crystal structure of the Lin28-interacting module of human terminal uridylyltransferase that regulates let-7 expression.
Nat Commun, 10, 2019
|
|
5WU5
 
 | | Crystal structure of apo human Tut1, form III | | Descriptor: | Speckle targeted PIP5K1A-regulated poly(A) polymerase | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2016-12-16 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.404 Å) | | Cite: | Crystal structures of U6 snRNA-specific terminal uridylyltransferase
Nat Commun, 8, 2017
|
|
5WU2
 
 | |
5WU6
 
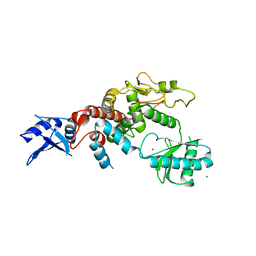 | | Crystal structure of apo human Tut1, form IV | | Descriptor: | MAGNESIUM ION, Speckle targeted PIP5K1A-regulated poly(A) polymerase | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2016-12-16 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.209 Å) | | Cite: | Crystal structures of U6 snRNA-specific terminal uridylyltransferase
Nat Commun, 8, 2017
|
|
5WU4
 
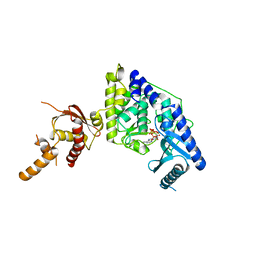 | |
5WU3
 
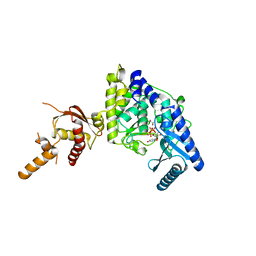 | | Crystal structure of human Tut1 bound with MgUTP, form II | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2016-12-16 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Crystal structures of U6 snRNA-specific terminal uridylyltransferase
Nat Commun, 8, 2017
|
|
5WU1
 
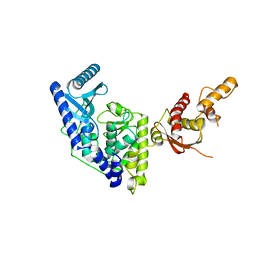 | | Crystal structure of apo human Tut1, form I | | Descriptor: | CHLORIDE ION, Speckle targeted PIP5K1A-regulated poly(A) polymerase | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2016-12-16 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of U6 snRNA-specific terminal uridylyltransferase
Nat Commun, 8, 2017
|
|
3VNV
 
 | | Complex structure of viral RNA polymerase II | | Descriptor: | 3'-DEOXY-CYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, Elongation factor Ts, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2012-01-18 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Mechanism for template-independent terminal adenylation activity of Q beta replicase
Structure, 20, 2012
|
|
3VNU
 
 | | Complex structure of viral RNA polymerase I | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Elongation factor Ts, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2012-01-18 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mechanism for template-independent terminal adenylation activity of Q beta replicase
Structure, 20, 2012
|
|
3WFR
 
 | | tRNA processing enzyme complex 2 | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Yamashita, S, Takeshita, D, Tomita, K. | | Deposit date: | 2013-07-23 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Translocation and rotation of tRNA during template-independent RNA polymerization by tRNA nucleotidyltransferase
Structure, 22, 2014
|
|
3WFO
 
 | | tRNA processing enzyme (apo form 1) | | Descriptor: | Poly A polymerase, SULFATE ION | | Authors: | Yamashita, S, Takeshita, D, Tomita, K. | | Deposit date: | 2013-07-23 | | Release date: | 2014-01-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Translocation and rotation of tRNA during template-independent RNA polymerization by tRNA nucleotidyltransferase
Structure, 22, 2014
|
|
3WFP
 
 | | tRNA processing enzyme (apo form 2) | | Descriptor: | Poly A polymerase, SULFATE ION | | Authors: | Yamashita, S, Takeshita, D, Tomita, K. | | Deposit date: | 2013-07-23 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.005 Å) | | Cite: | Translocation and rotation of tRNA during template-independent RNA polymerization by tRNA nucleotidyltransferase
Structure, 22, 2014
|
|
3WFS
 
 | | tRNA processing enzyme complex 3 | | Descriptor: | Poly A polymerase, RNA (74-MER), SULFATE ION | | Authors: | Yamashita, S, Takeshita, D, Tomita, K. | | Deposit date: | 2013-07-23 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.311 Å) | | Cite: | Translocation and rotation of tRNA during template-independent RNA polymerization by tRNA nucleotidyltransferase
Structure, 22, 2014
|
|
3WFQ
 
 | | tRNA processing enzyme complex 1 | | Descriptor: | Poly A polymerase, RNA (73-MER) | | Authors: | Yamashita, S, Takeshita, D, Tomita, K. | | Deposit date: | 2013-07-23 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.619 Å) | | Cite: | Translocation and rotation of tRNA during template-independent RNA polymerization by tRNA nucleotidyltransferase
Structure, 22, 2014
|
|
2Z3K
 
 | | complex structure of LF-transferase and rAF | | Descriptor: | 2-(6-AMINO-OCTAHYDRO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, D(-)-TARTARIC ACID, Leucyl/phenylalanyl-tRNA-protein transferase, ... | | Authors: | Watanabe, K, Toh, Y, Tomita, K. | | Deposit date: | 2007-06-04 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Protein-based peptide-bond formation by aminoacyl-tRNA protein transferase
Nature, 449, 2007
|
|
2Z3M
 
 | | complex structure of LF-transferase and dAF | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, D(-)-TARTARIC ACID, Leucyl/phenylalanyl-tRNA-protein transferase, ... | | Authors: | Watanabe, K, Toh, Y, Tomita, K. | | Deposit date: | 2007-06-04 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Protein-based peptide-bond formation by aminoacyl-tRNA protein transferase
Nature, 449, 2007
|
|
2Z3L
 
 | | complex structure of LF-transferase and peptide A | | Descriptor: | D(-)-TARTARIC ACID, Leucyl/phenylalanyl-tRNA-protein transferase, peptide (PHE)(ARG)(TYR)(LEU)(GLY) | | Authors: | Watanabe, K, Toh, Y, Tomita, K. | | Deposit date: | 2007-06-04 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Protein-based peptide-bond formation by aminoacyl-tRNA protein transferase
Nature, 449, 2007
|
|
2Z3P
 
 | |
2Z3O
 
 | |
2Z3N
 
 | | complex structure of LF-transferase and peptide B | | Descriptor: | D(-)-TARTARIC ACID, Leucyl/phenylalanyl-tRNA-protein transferase, peptide (PHE)(ARG)(TYR)(LEU)(GLY) | | Authors: | Watanabe, K, Toh, Y, Tomita, K. | | Deposit date: | 2007-06-04 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protein-based peptide-bond formation by aminoacyl-tRNA protein transferase
Nature, 449, 2007
|
|
3AQK
 
 | |
3AQN
 
 | |
3AQL
 
 | |
